Simon Myers
@simon_r_myers
Followers
603
Following
43
Media
0
Statuses
65
Joined September 2018
10) Biggest caveats: for technical reasons we only look at quantitative traits, not e.g. disease presence/absence. We studied only UK-born people: genetic effects might differ more strongly between individuals from different countries, for example.
0
0
4
9) We also map fine-scale population differences across regions of the UK, and show that for traits which themselves vary geographically, these remove false positives and false negatives in GWAS which occur using leading present-day approaches
1
0
2
8) Again though, there may be good news. If this is the main problem, there is hope of narrowing down hidden causal variants by combining information across populations, leveraging that common effects operate, to make "portable" scores
1
0
1
7) .....and in other ancestries causal mutations might either not be found, or be poorly captured (i.e. tagged) by mutations that do this well in European-ancestry people. This is a well-known problem.
1
0
0
6) So why the poor performance of the initial polygenic scores, in non-Europeans?? This may almost entirely reflect either that these scores are made in individuals of European ancestry, who form the majority of the UK Biobank cohort.....
1
0
2
5) This is good news! It offers the hope that, IF causal mutations are shared between two populations, they are likely to work the same way. This makes biomedical application of genetic findings across ethnicities more straightforward.
1
0
3
4) This contrasts with some (though not all) previous studies, which conclude that effect sizes DO differ strongly across groups. It means that although genes may strongly interact with e.g. the environment, these interactions are mainly shared between populations.
1
0
0
3) Combining with other recent studies e.g. https://t.co/rg6EdEqSv5 , we can conclude that a mutation increasing e.g. cholesterol or blood pressure in a European-ancestry individual has a near-identical effect in someone of African-ancestry, at least if both are UK-born
nature.com
Nature Genetics - This analysis of individuals of admixed genetic ancestries suggests that complex trait causal variant effect sizes are, by and large, similar across ancestries, and discusses the...
1
0
0
2) However, when we looked *only* at regions of European ancestry, within the genomes of mainly African-ancestry individuals, scores from these regions predicted traits just as well as if they had occurred in ~100% European-ancestry individuals.
1
0
0
Thread: 1) We constructed polygenic scores to predict 29 quantitative phenotypes in UK Biobank individuals of different ancestries. As previous studies, we found that these scores predicted well in European-ancestry individuals, but very poorly in e.g. African-ancestry people
1
0
0
New manuscript on biorxiv, led by the wonderful Sile Hu! We show underlying phenotypic effects are highly conserved across people from different ethnic groups living in the UK, despite the apparent non-portability of polygenic predictions. https://t.co/BcFZzBEGAF
biorxiv.org
An understanding of genetic differences between populations is essential for avoiding confounding in genome-wide association studies (GWAS) and understanding the evolution of human traits. Polygenic...
2
20
79
We are hiring! We're seeking a senior experimental post-doc to work on meiosis, gene regulation and single-cell data, deadline 28th April:
0
5
3
In-person registration for Probabilistic Modelling in Genomics 28th-30th March, Oxford has been extended a few days until Friday - last few rooms left! Online registration will stay open. #ProbGen22
probgen22.github.io
Probabilistic Modelling in Genomics 2022, 28-30th March, Oxford.
0
6
9
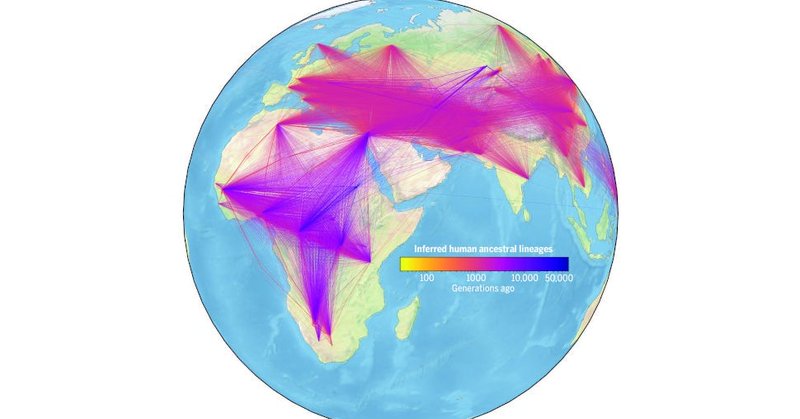
Massive New ‘Human Family Tree’ Includes 27 Million Ancestors
gizmodo.com
The expansive genealogy contains critical events from our distant past, including the first migrations out of Africa.
2
13
36
Fantastic list of plenary speakers for #ProbGen22!
Excited to announce our stellar plenary speaker lineup for #ProbGen22: @marakat, @erikmvolz and @Graham_Coop. Register now to avoid disappoinment!
0
0
4
Pier Palamara’s research group at @OxfordStats and @HumanGeneticsOx is recruiting an ERC-funded postdoctoral research associate. Large-scale genealogical inference for population genetics and complex trait analysis. More info:
0
21
16
Registration is open for Probabilistic Modelling in Genomics 28th-30th March, Oxford. Please RT! #ProbGen22
probgen22.github.io
Probabilistic Modelling in Genomics 2022, 28-30th March, Oxford.
1
14
21
Save the date for #ProbGen22: 28th-30th March! Probabilistic Modelling in Genomics is in Oxford, jointly hosted by @bdi_oxford, @HumanGeneticsOx and @OxfordStats. Three day hybrid event, US East coast friendly scheduling. More details to follow soon. Please RT!
0
19
42
📢Three exciting jobs! Florence Nightingale Bicentennial Fellow in Statistics, Probability or a related subject (x2) and Glasstone Research Fellowship in Science, both for researchers at an early stage of their career. More details:
1
8
12
Help me raise £300 to help raise £100,000 for a new track and field area for Bicester children's athletics club, and I will run London marathon in return. Please #donate on @justgiving and RT. Thanks!
justgiving.com
Help Simon Myers raise money to support a great cause
0
1
0