Potestio Lab
@r_potestio
Followers
634
Following
3K
Media
154
Statuses
518
Research team led by Raffaello Potestio at the University of Trento, Italy. Multi-scale modelling, computational biophysics, statistical mechanics, and pizza.
Trento, Trentino-Alto Adige
Joined November 2016
Biophysics, Stat Mech and Machine Learning will meet in Trento from July 7th to 11th, 2025 in our StatPhys29 Satellite Workshop "Molecular biophysics at the transition state: from statistical mechanics to AI": Co-organized with @ai_ngrosso.
0
2
4
This research is the result of a collaborative effort among the teams at the University of Trento, INFN-TIFPA, and the Donders Institute, and we are grateful for the support of our colleagues and institutions. #MachineLearning #DeepLearning #NeuralNetworks #StatPhysics.
0
0
0
🥳 We are pleased to announce the publication of our paper “Density of States in Neural Networks: An In-Depth Exploration of Learning in Parameter Space” in Trans. on Machine Learning Research. @MeleMargherita_ @ai_ngrosso @UniTrento @INFN_ @DondersInst.
openreview.net
Learning in neural networks critically hinges on the intricate geometry of the loss landscape associated with a given task. Traditionally, most research has focused on finding specific weight...
1
1
2
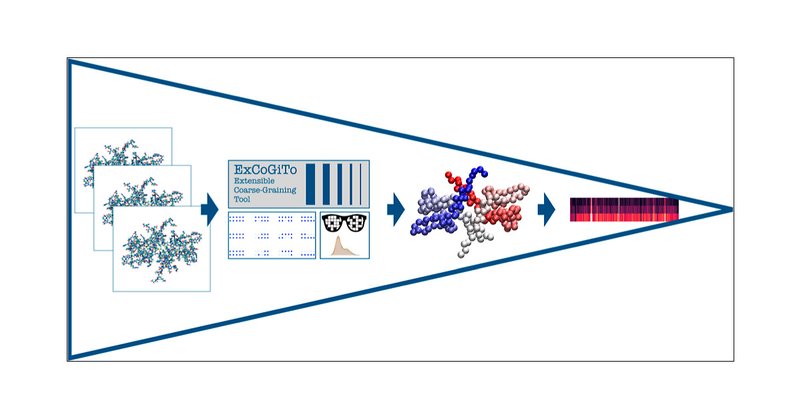
The MEOW approach, implemented in the EXCOGITO software suite, attributes to atoms and residues a relevance score based on their dynamical and energetic properties, and allows one to rationalise their functional behaviour.
pubs.acs.org
Bottom-up coarse-grained (CG) models proved to be essential to complement and sometimes even replace all-atom representations of soft matter systems and biological macromolecules. The development of...
1
0
0
Our PhD student Camilla Spreti @Camilla_Spreti just gave a talk at the yearly PhD Workshop of the Physics Dept. of the University of Trento @UniTrento, illustrating her research on enhanced sampling of small stat mech systems. Excellent job! 🥳.
0
2
4