Petr Šulc
@petr_sulc
Followers
1K
Following
4K
Media
117
Statuses
598
Associate Professor at ASU and ERC PI at TUM. We use computer simulations and experiments to play with DNA and RNA in biology and nanotechnology
Tempe, AZ; Munich, DE;
Joined March 2014
For the newest update for our group research, please follow our new profile on bluesky:
0
0
0
Sharing a paper below in @CellGenomics that took us literally 10 years+ (!) to write on the topics of 👇👇👇 * How, exactly, do we quantify viral mimicry - the expression of molecular features usually associated with nonself viruses by our own "self" cells? * Why do we see
0
10
26
Navrhuje DNA a RNA nanostruktury – stavebnice budoucnosti pro diagnostiku, léky i nové materiály. Laureát Ceny Neuron 2024, profesor na Arizona State a TU Mnichov, autor 70+ publikací včetně Science a Nature Nanotechnology. 💡 Poznejte Petra Šulce: 👉 https://t.co/ywfbsRYKeb
0
1
5
Many thanks to @NadaceNeuron for this award, and my students and collaborators who made all this work that was recognized possible.
Big congrats to SMS's Associate Professor Petr Sulc for winning the Neuron Foundation "Promising Scientist in Physics" prize! 🏆 This prestigious Czech award celebrates his contributions to nucleic acids modeling and bionanotechnology. 🎉👏
1
2
30
Our new article with J Russo, D. Pinto and F. Sciortino out in @acsnano : We use our #SATAssembly method to design particles that assemble into 3D quasicrystal clusters, paving a way to their experimental realization with DNA nanostructures:
pubs.acs.org
One of the frontiers of nanotechnology is advancing beyond the periodic self-assembly of materials. Icosahedral quasicrystals, aperiodic in all directions, represent one of the most challenging...
0
1
12
For more updates about our lab's research, we are switching to our bluesky profile:
0
0
2
Our latest work, where we use DCA as a generative model for peptide binders in collaboration with our colleagues from @CaFoscari :
Combination of Coevolutionary Information and Supervised Learning Enables Generation of Cyclic Peptide Inhibitors with Enhanced Potency from a Small Data Set #machinelearning #compchem
1
0
0
Combination of Coevolutionary Information and Supervised Learning Enables Generation of Cyclic Peptide Inhibitors with Enhanced Potency from a Small Data Set #machinelearning #compchem
pubs.acs.org
Computational generation of cyclic peptide inhibitors using machine learning models requires large size training data sets often difficult to generate experimentally. Here we demonstrated that...
0
4
21
Using @ox_DNA simulations and experiments, @petr_sulc lab shows that adding functionalization (e.g. proteins) onto a DNA origami causes entropy-driven bending, with implications for nanostructure designs Read it here 🔗 https://t.co/GE2VkVhx2R
1
3
10
.@petr_sulc, @progolab et al. demonstrate how the presence of single- & double-stranded overhangs drive entropy-induced conformational changes in 2D #DNAOrigami #nanostructures. #oxDNA @ASUBiodesign @ASU @TUM_STS @TU_Muenchen Read: https://t.co/sS6TVuQlsT
0
7
16
5/5: We also confirmed the effect when proteins are attached to DNA origami surface, using both experiments and simulations (with ANM-oxDNA model). The article is out at: https://t.co/rsmbWcuHyF Excellent work by my group: M. Sample, @progolab , T. Diep and H. Liu
0
0
5
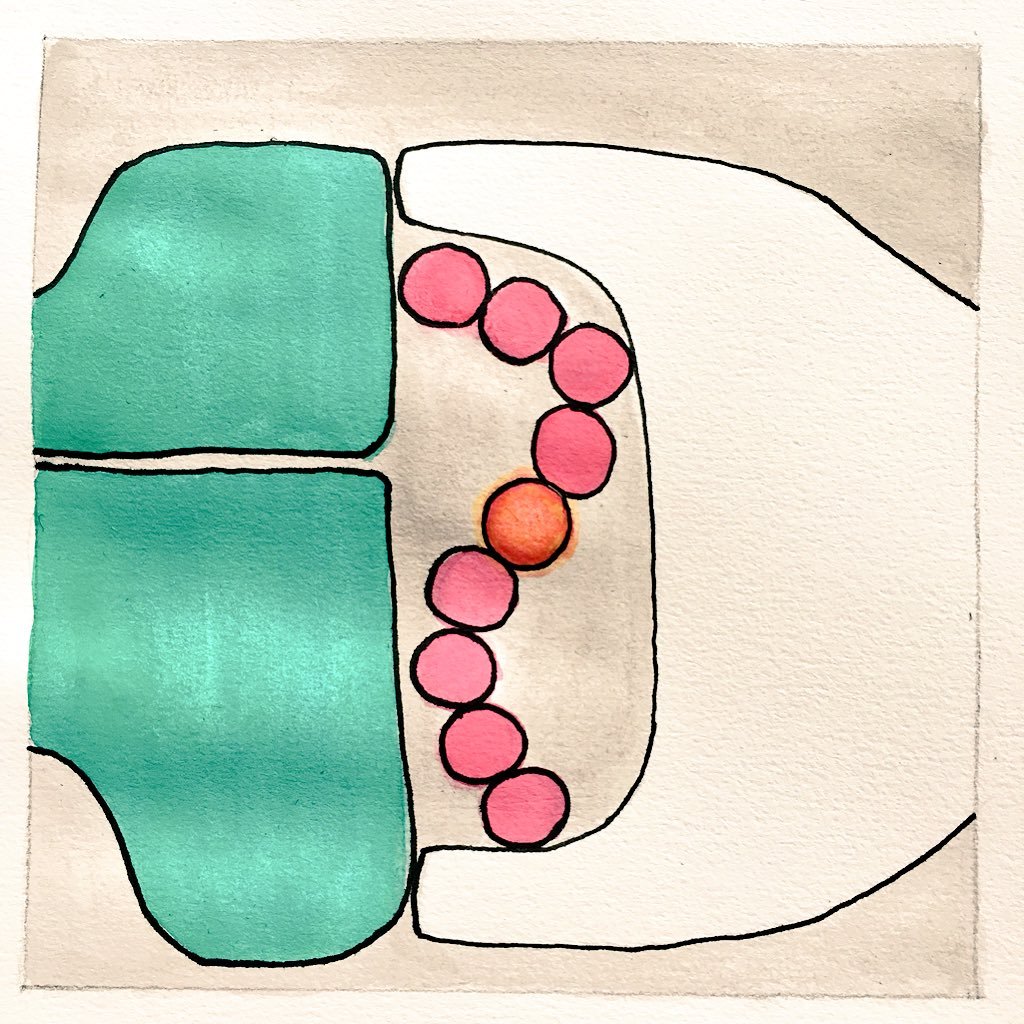
4/5: We then verified the predictions experimentally, both with attached DNA overhangs: 2D sheets with induced bending preferentially form tubes, while the non-functionalized ones are flatter and tend to form sheets.
1
0
0
3/5: The bent structure gives larger conformational space to the individual overhangs, making the bent conformation (ie small edge-to-edge distance on the tile) more likely. We used @ox_DNA to sample the free-energy landscape, assigning probabilities to different conformations.
1
0
0
2/5:Attaching overhangs to the 2D origami causes its conformational space to change, leading to the more curved conformations being more likely. This can have important implications for the desired function (e.g precise protein patterning).The reason for this bending is entropic
1
0
0
Trouble with too much hair? In our new paper, just out in @acsnano , we investigate what we call the "hairygami" effect: the entropy-induced bending of flexible DNA origami sheet at nanoscale when you functionalize it e.g. with ssDNA overhangs or attached proteins: 🧵1/5 ⬇️:
1
3
24
We are grateful for our new NSF POSE grant that allows us to expand and establish oxDNA open source ecosystem. Do you use (or want to) @ox_DNA , are there any features that you miss, any bottlenecks? We want to hear from you so that we can make the tools better, please DM me!
0
4
20
Excited to join the @foresightinst workshop on nanotechnology!
We invite you to apply to not only one but two events to advance bottlenecks to nanotechnology development: 1. Molecular Systems Engineering Platform Launch One of the major bottlenecks holding back progress on nanotechnology is better software and simulation tools—until now.
0
0
6
We invite you to apply to not only one but two events to advance bottlenecks to nanotechnology development: 1. Molecular Systems Engineering Platform Launch One of the major bottlenecks holding back progress on nanotechnology is better software and simulation tools—until now.
1
4
7
A new tutorial by our student Sarah on using @ox_DNA ecosystem to simulate DNA origami: https://t.co/5R72qlaGBp No coding needed, just web browser and your starting design file (cadnano or oxdna file format). Article also accompanied by video tutorial: https://t.co/bEOhjgKDzp
2
11
59
Our collaboration with Rief and Simmel labs is now out in @NatureComms : https://t.co/lpiuYf7Bt7 A single-molecule study of strand invasion reactions: DNA invading DNA, RNA invading RNA, as well as DNA/RNA hybrids, accompanied by our @ox_DNA simulations
nature.com
Nature Communications - This study uses single molecule mechanical experiments and computer simulations to measure the speed by which an invading DNA or RNA strand displaces a bound strand from a...
0
2
28