Matthew Solomonson
@mattsolomonson
Followers
418
Following
548
Media
11
Statuses
178
Associate Director, Genomic Data Visualization @ The Broad Institute of MIT and Harvard
Cambridge, MA
Joined January 2011
Super excited to release a web application for browsing association results for 3,700 phenotypes across 281,850 individuals from the UK Biobank. The site features plots, tables, and controls for exploring billions of associations.
7
200
553
RT @gnomad_project: The #gnomAD team is proud to announce the release of gnomAD v4! The v4 dataset includes 730,947 exomes & 76,215 genomes….
0
255
0
RT @gnomad_project: The #gnomAD browser now includes variant co-occurrence counts by gene. We hope this will help with interpretation of co….
0
51
0
RT @ksamocha: New feature on gnomAD! For pairs of rare variants, we predicted the number of individuals in gnomAD with these variants in ci….
0
37
0
RT @HeidiRehm: @globalbiodata We are delighted that @ClinGenResource and @gnomad_project were chosen as GCBRs! Thank you to the efforts of….
0
3
0
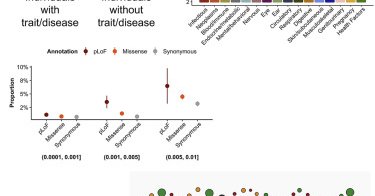
RT @konradjk: Our paper on rare variant associations in the UK Biobank is out in Cell Genomics today!
cell.com
Karczewski et al. generated a massive-scale association dataset between rare genetic mutations and thousands of diseases and traits and released these data in the Genebass browser. They quantify the...
0
79
0
RT @konradjk: Our preprint on rare variant associations in the UK Biobank exomes has now been updated to its final release of 450K exomes:….
0
42
0
RT @dgmacarthur: Amen to this whole article, but especially this paragraph. If we want to do robust, reproducible, large-scale science, aca….
0
28
0
RT @HeidiRehm: We're excited to recruit a new software engineer for our @gnomad_project team at the @broadinstitute working on the https://….
0
19
0
RT @gnomad_project: Our browser now includes short tandem repeat (STR) calls for 59 disease associated loci generated by running ExpansionH….
0
29
0
RT @ksamocha: Great to see the #AlphaFold predictions incorporated into the @decipher_wtsi browser so quickly! Cool to be able to see patie….
0
11
0
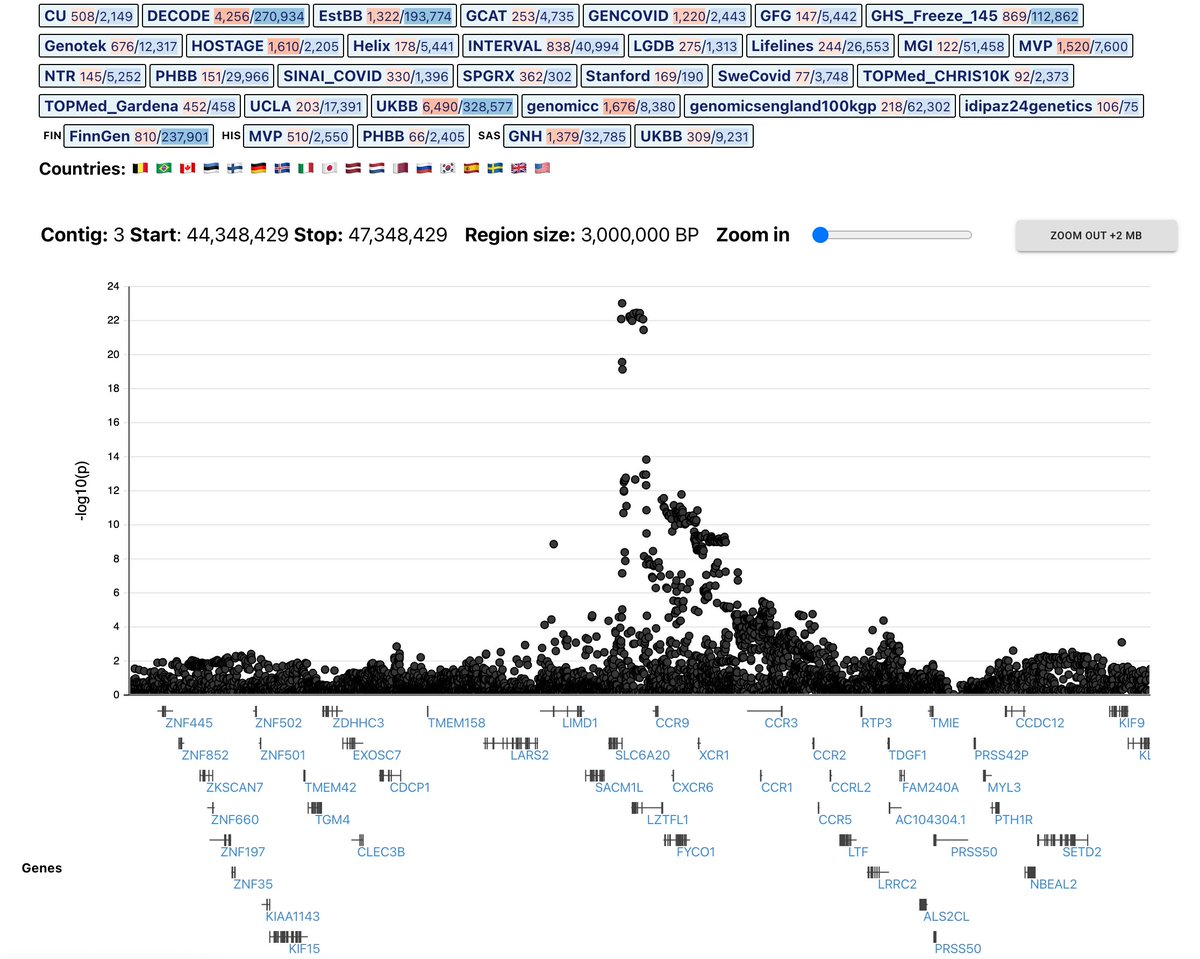
Excited to share this short YouTube walk-through of Genebass, produced for the @uk_biobank Scientific Conference #UKBSC21.
youtube.com
A walkthrough of the Genebass resource (https://genebass.org/).
0
0
3
RT @dgmacarthur: After nearly a decade (!) of iterative development on the seqr platform for management and analysis of rare disease genomi….
medrxiv.org
Exome and genome sequencing have become the tools of choice for rare disease diagnosis, leading to large amounts of data available for analyses. To identify causal variants in these datasets,...
0
34
0
RT @mbeisen: If I could snap my fingers and change everything, here is what I would do:. 1) Have all publishing happen on a single, consoli….
0
148
0
RT @CovidCg: Today is s 1-year anniversary 🎉🎋🥳🎂. We have just released a new Lineage Reports feature on the site to….
0
10
0
RT @d0choa: Systematically overlaying likely pathogenic variants over @DeepMind #AlphaFold predicted structures provides a unique opportuni….
0
53
0
RT @Ayjchan: A picture of SARS-CoV-2 variants in the USA. Plotting the lineage percentages of new sequences each day over the past 6 mont….
0
46
0
RT @DeepMind: Today with @emblebi, we're launching the #AlphaFold Protein Structure Database, which offers the most complete and accurate p….
0
3K
0
@huy_dev set up an impressive mechanism for enabling ~85 translators (!) to translate blog posts in >30 languages. The posts are drafted by @mkveerpn and @bnwolford’s scientific communication team. Each translation gets its own sharable deployment & preview.
1
1
2