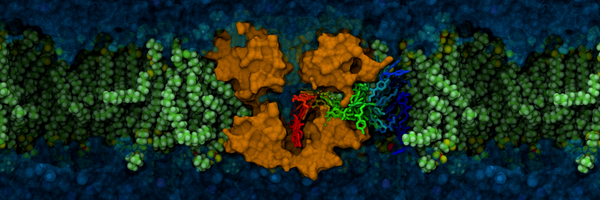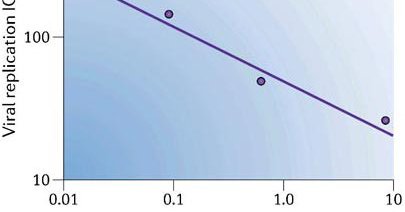
lewis martin
@lewischewis
Followers
1K
Following
13K
Media
94
Statuses
600
Blog: Use RDKitJS to search for similar ligands in the PDB in the browser. https://t.co/hjIdVPwjWz
1
4
40
spmat.A... no spmat.asnumpy()... spmat.numpy()... spmat.array().... spmat.asarray()... spmat.toarray() ah
0
0
4
new blog: installing and running BioEmu on Modal for the gpu poor:
ljmartin.github.io
for GPU poor
0
1
10
Running solubility predictions couldn't be easier! With Rowan's latest release, you can go from drawing a molecule to viewing predicted temperature- and solvent-dependent solubility in <2 minutes. (Video sped up 2x because we respect your time)
3
27
178
RDKit is the standard software for cheminformatics, but RDKit doesn’t integrate naturally with high-accuracy chemistry simulation tools. Starting from RDKit, it's non-trivial to simulate pKa, optimize conformers w/ ML, score tautomers, &c. There are many posts like this:
2
6
60
We all know about cell potency dropoff due to permeability, efflux, or plasma binding. This adds another angle: diffusion inside the cytoplasm is 10X slower than water, so your effective Kd in the cell is worse! Off-rate remains the same, but fast on-rate requires fast diffusion
New Blog: A protein takes 0.01 s to traverse E. coli's diameter. The same protein takes 4 min to move 1 mm and >6 hours to move 1 cm. The farther a molecule must move, the less likely it will get there. This is partly why cells are very tiny — it makes collisions more likely.
0
0
14
Unlock the mystery of entropy with my latest #StatisticalMechanics lecture now on YouTube! 🌪️📷 We delve into how entropy S relates to disorder W, revealing that S = kln(W), k being Boltzmann's constant. Thanks to Stirling, this simplifies to S = kln(N) for N equally likely
0
3
38
New preprint! With @JosephJGair1 + the Gair lab @MSUChem, we built a set of strained conformers for benchmarking DFT functionals and NNPs. Our new benchmark set, "Wiggle150", is substantially harder than previous conformational benchmarks, with an avg ∆E of 103 kcal/mol.
4
25
118
then that it seems to match sota RBFE tools at <100X the compute is the cherry on top
0
1
6
it's like a mechanical bull for calculating off-rates. And Copeland teaches us that off-rates are what you need to optimize a series, since on-rate has an upper ceiling - the diffusion limit in solution
nature.com
Nature Reviews Drug Discovery - Pharmacological activity depends on the binding of drugs to their targets. Copelandet al. provide a perspective on the importance of residence time for lead...
2
1
7
this paper is sooo good. boils down to 'what if I just simulate at 1000 degrees and see how long each ligand can hold on for?' and it works
3
2
51
After long days spent on isosurface visualization, sparse data storage, and the like ... @RowanSci's orbitals workflow is live! Molecular orbitals, computed with QM, provide a picture of the location of electrons, yielding insights about bonding, excited states, and reactivity.
3
4
27
Now upgraded to a fully-fledged @modal_labs example! Fold proteins at scale, without getting a second PhD in Kubernetes. https://t.co/7dhvZIjqRX
modal.com
In biology, function follows form quite literally: the physical shapes of proteins dictate their behavior. Measuring those shapes directly is difficult and first-principles physical simulation...
1
2
14
Predicting multiple conformations of ligand binding sites in proteins suggests that AlphaFold2 may remember too much @PNASNews • This study investigates AlphaFold2’s ability to generate conformational ensembles of protein binding sites, revealing limitations tied to its
0
17
84
attn GPU-poor: here's how to run Chai-1 on a Modal instance
ljmartin.github.io
A100s for all
Chai-1 has always been available for commercial use via our server. Today, we're also making Chai-1(r) code and weights available under an Apache 2.0 license, which permits broad commercial use. https://t.co/EFz6edbreN
0
2
28
Blog: an equation to combine the Kd of two ligand species, when you know their populations, using kinetic models with scipy odeint
1
0
4
blog post: I found this pretty memory hungry, so here's how to run Boltz-1 on A100's using Modal:
ljmartin.github.io
A100s for all
Thrilled to announce Boltz-1, the first open-source and commercially available model to achieve AlphaFold3-level accuracy on biomolecular structure prediction! An exciting collaboration with @jeremyWohlwend, @pas_saro and an amazing team at MIT and Genesis Therapeutics. A thread!
1
7
48
tip: if you save SDF blocks into a parquet file, you can write a multi-molecule SDF output with duckdb like: ``` .mode ascii .headers off -- make some table including 'sdf' col select sdf || '$$$$' || chr(10) from mytable; ```
0
0
3
We're launching individual subscriptions to @RowanSci! For $200/month, you can run >20 hours of calculations every week and gain access to advanced workflows (BDE + more soon). Here's our motivation for this change, what this means for existing users, and where we're going:
2
10
29
PocketVec is published in @NatureComms and I still can't believe it. Huge thanks to @jorba_guillem, @XavierBarril and @ptck72 !! https://t.co/3r1W6TIY5X
2
14
77