Jacob Schreiber
@jmschreiber91
Followers
5K
Following
7K
Media
500
Statuses
5K
Guest Scientist @impvienna, Board of Directors @NumFOCUS, incoming prof @UMassGCB. Prev, @StanfordMed @uwcse. Studying genomics, machine learning, and fruit.
Vienna, Austria
Joined March 2017
The more papers I read for a review article I'm writing about ML pitfalls in genomics, the more my faith is shaken in the results from papers that apply machine learning to methylation arrays. A salty thread. 1/.
21
220
825
RT @pkoo562: Our work on "Evaluating the representational power of pre-trained DNA language models for regulatory genomics" led by @AmberZq….
genomebiology.biomedcentral.com
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of...
0
42
0
RT @jmschreiber91: Today marks the end of en-JUNE-eering, the month where I focused mostly on the nitty gritty of improving genomics ML inf….
0
4
0
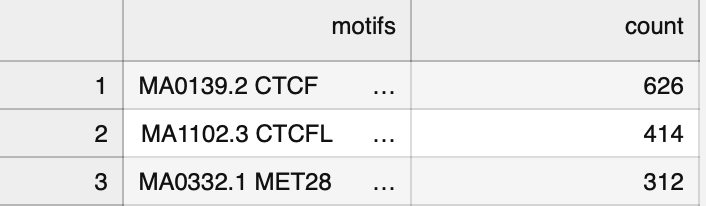
(5) A new tomtom-lite command-line tool that allows quick querying of motifs without needing to go to the Tomtom website.
Last week I shared tomtom-lite, a super fast re-implementation of Tomtom for annotating short genomic spans with the motifs they most resemble. Now, there's a convenient command line tool `ttl` that comes with the installation. You can get it with `pip instal memelite`.
0
0
3
(4) bpnet-lite: Load Chrom/BPNet models from the official TensorFlow repos into PyTorch for downstream tangermeme. integration Improved command-line tools + docs. Still concerns about perf of models trained from scratch -- will be resolved next version!.
Last week I released bpnet-lite v0.5.0. Chrom/BPNet are powerful models for understanding regulatory genomics from @anshulkundaje's group, and now it's way easier to go from raw data to trained models and analysis/results in PyTorch . Try it out with `pip install bpnet-lite`.
1
0
2
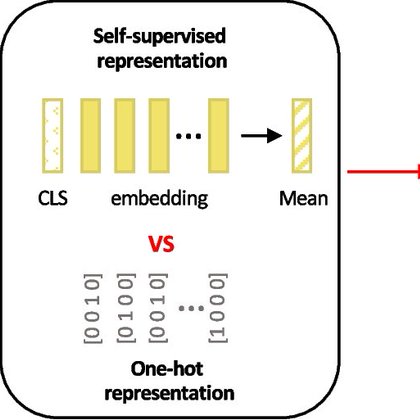
(3) tangermeme: significant quality-of-life improvements, fixing an issue with seqlet calling, plotting w/ annotations, and tomtom-lite integration across several functions.
github.com
Biological sequence analysis for the modern age. Contribute to jmschrei/tangermeme development by creating an account on GitHub.
Just released tangermeme v0.5.0! . tangermeme implements "everything-but-the-model" for genomic ML Train your model your way using your code-base (or load someone else's model), and tangermeme handles the discovery + design with it. Try it out with `pip install tangermeme`.
1
0
1
(2) bam2bw: a simple utility that allows you to go from BAMs -> un/stranded bigWigs without intermediary bedGraph files. Way less memory, disk, and hassle. Now extended to work on fragment files and .tsv/.gz, and depth normalize.
github.com
A command-line tool for reading SAM/BAM files and converted them directly to bigwig files. - jmschrei/bam2bw
Released a new version of my little data processing tool, `bam2bw`. As you might expect, it goes directly from BAM to un/stranded bigwig(s), but now also supports fragment.tsv files. No more intermediary sorting and bedGraph steps! Way faster + less disk than before.
4
0
4
(1) tomtom-lite: a significantly faster implementation of the original tomtom algorithm that can be over 1000x faster. Now built-in to a variety of my other tools.
github.com
A lightweight reimplementation of some of the algorithms in the MEME suite in Python. - jmschrei/memesuite-lite
I wrote a quick application note on Tomtom-lite, a Python implementation of the Tomtom algorithm for comparing PWMs against each other. This implementation can be 10-1000x faster and, as a Python function, can be integrated into your workflows easier.
1
0
2
This evaluation of DNA design methods is very well written. If you're interested in the field, you should def take a look. Also, glad to see Ledidi performing so well!.
biorxiv.org
One outstanding open problem with high therapeutic value is how to design nucleic acid sequences with specific properties. Even just the 5’ UTR sequence admits 2 × 10120 possibilities, making...
0
8
54
In vivo mapping of mutagenesis sensitivity of human enhancers.
nature.com
Nature - Human enhancers contain a high density of sequence features that are required for their normal in vivo function.
1
0
13