Jérome Eberhardt
@je_eberhardt
Followers
163
Following
405
Media
30
Statuses
455
Postdoc at @biozentrum (unibas), lazy python ninja (https://t.co/uVqpjgdMru) and outside the wrong box thinker.
Joined April 2009
Thanks to everyone who joined us last week! You can now watch the recording of the webinar:
We’re hosting @peter_skrinjar for a webinar to discuss his latest paper “Have protein co-folding methods moved beyond memorization?” 🚀 Peter will discuss the limitations of current co-folding models, the challenges of memorization in ligand pose prediction, and what is needed
0
1
3
Join us tomorrow to hear @peter_skrinjar talk about 🌹Runs N' Poses🌹! Curious to hear your thoughts on how to keep up with benchmarking in this field. More info:
We’re hosting @peter_skrinjar for a webinar to discuss his latest paper “Have protein co-folding methods moved beyond memorization?” 🚀 Peter will discuss the limitations of current co-folding models, the challenges of memorization in ligand pose prediction, and what is needed
0
2
3
Watch the Webinar: Python Scripting for Molecular Docki This virtual course introduces life scientists to the power and flexibility of python scripting https://t.co/2ndCAXL0po
rcsb.org
This virtual course introduces life scientists to the power and flexibility of Python Scripting for Molecular Docking
1
74
299
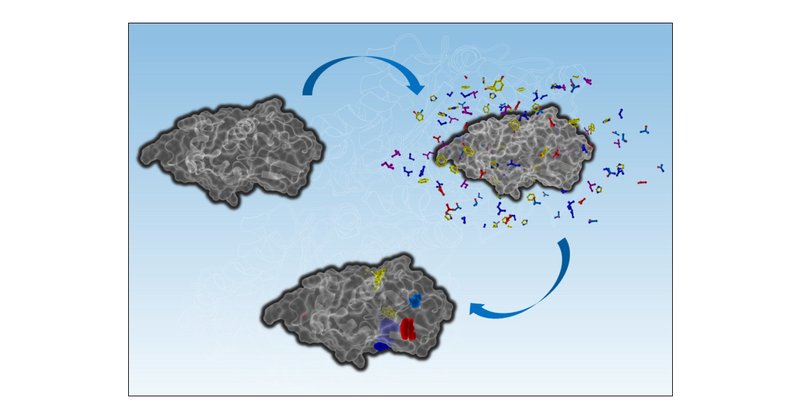
Are you struggling to model your ligand into x-ray crystallography or cryo-EM density? Trying to dock a novel ligand into a well-studied target but just cannot get the pharmacophore position correct? Running a virtual screening and want to optimize your hit rate? (1/4)
3
8
32
If you’re interested in fast sampling and flexible and automated creation of cosolvent systems with @openmm_toolkit check this out! Huge thanks to the team @je_eberhardt @diogo_stmart @manuco17 @moonii1020 @Joe_r_loeffler @ForliLab
https://t.co/EJhjQuW2En ⚛️🧬
pubs.acs.org
Cosolvent molecular dynamics (MDs) are an increasingly popular form of simulations where small molecule cosolvents are added to water-solvated protein systems. These simulations can perform diverse...
1
11
23
Buried under a pile of docking data and not sure where to go next? Ringtail 2 will help you organize, filter, and visualize docking data via an easy-to-use command line tool, or integrated in your #python scripts using the brand new API! #autodockgpu #autodockvina
1
10
37
Do you know Python and have experience with RDKit? Do you want to work in an exciting and multidisciplinary environment?The AutoDock project is hiring a programmer, apply here!
1
13
36
The bottleneck for a protein-ligand complex (PLI) prediction breakthrough isn't a lack of ML innovation, but accessible data and evaluation to show which ideas work and how to improve them. We (@TorstenSchwede, @vant_ai, @NVIDIA, @MIT_CSAIL) present PLINDER 🧵(1/n)
6
60
248
Postdoc in Computational Structural Biology - Evolutionary-scale interpretation of protein functions in human gut microbiome (80-100%) https://t.co/9xnGSrv9Cu
@biozentrum @UniBasel_en
#Biozentrum #Basel #research #science #joboffer #postdoc
5
15
16
… and with small-molecules!
CASP16 Call for Targets Submit your targets to Critical Assessment of protein Structure Prediction, including structures determined by multiple experimental methods (cryoEM, NMR, X-ray crystallography, SAXS, cross-link) https://t.co/aFKOLCFbMS
0
0
2
1
0
0
Parce que bon, tout l’avantage d’EWLD c’est qu’il est peu cher et capitalisant !
1
0
1
From January 2025, Torsten Schwede will be President of the SNSF Research Council. The renowned bioinformatician and current Vice President for Research at the University of Basel will support the restructuring of the Research Council already in 2024. https://t.co/voxCyuKZyb
snf.ch
The Swiss National Science Foundation (SNSF) funds excellent research at universities and other institutions.
2
10
49
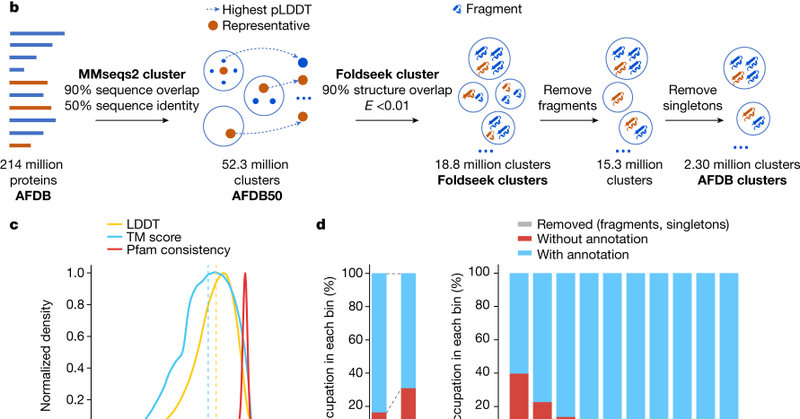
So happy to see our work going live together with that of @pedrobeltrao and @thesteinegger today @nature! @biozentrum @UniBasel @ISBSIB @SWISS_MODEL link: https://t.co/nyXCNgxAHg link:
nature.com
Nature - The novel Foldseek clustering algorithm defines 2.30 million clusters of AlphaFold structures, identifying remote structural similarity of human immune-related proteins in prokaryotic...
12
71
189
Welcome to our #BiozentrumResearchSummer students! 13 international Bachelor students are currently experiencing hands-on research in our @biozentrum labs for up to 9 weeks. Enjoy an exciting summer as junior scientists in cutting-edge research https://t.co/D4XbrEKhVs
@unibas_en
1
5
25
Facilitating the access and reuse of computer-predicted protein structures with #ModelArchive. An initiative led by SIB Group Leaders has been awarded funding by @CH_universities to expand #OpenResearchData. Find out more: https://t.co/UCZG009tik
#deeplearning #AI
0
7
7
Time for a pip-update and/or pull some code from GitHub! We have a new Vina release v.1.2.4 with a few bug fixes and improvements. Release notes and binaries to download are available here: https://t.co/JGSZkNe1iD
0
6
19
The superb work of @je_eberhardt is finally out! @JCIM_JCTC Check out WaterKit, a GIST-based method written in Python for calculating thermodynamic properties of waters on protein surfaces with results comparable to MD simulations https://t.co/4t7F1PRSAi
3
28
69
You know Python, RDKit, and want to work in an exciting multidisciplinary environment? We're looking for programmers to design our next-gen molecular modeling tools. Apply here! Research Programmer III https://t.co/aVNdoVPzN3 Research Programmer IV https://t.co/acmoA3FHVj
1
9
17