Iker Rivas-González
@irg_bio
Followers
423
Following
485
Media
78
Statuses
208
Postdoctoral Researcher at @MPI_EVA_Leipzig ~ Primate evolution 🙈 ~ DataViz enthusiast 📈 ~ amateur crocheter 🧶 ~ they/them 🏳️🌈
Aarhus, Denmark
Joined March 2020
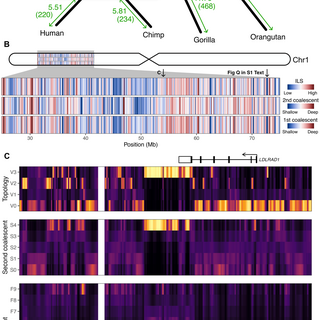
I am delighted to present TRAILS, a new hidden Markov model that.- reconstructs the speciation process (ancestral Ne and speciation times).- infers the multi-species ancestral recombination graph (ARG).Stay here for a brief summary of the paper! (1/17).
journals.plos.org
Author summary DNA sequences can be compared to reconstruct the evolutionary history of different species. While the ancestral history is usually represented by a single phylogenetic tree, speciation...
5
58
156
I cannot overstate how much of an incredible opportunity this is! The microbiome work of @DebrayReena and @jtung5 is cutting-edge and extraordinary. Come join our department here in beautiful Leipzig!.
The Social Microbiome Group is coming to @MPI_EVA_Leipzig and we are recruiting at multiple levels! You will work closely with me and Dr. Jenny Tung @jtung5. Apply by Sep 1 with a cover letter and CV. Please share 🔁
0
2
3
RT @PLOSGenetics: Researchers from @AarhusUni_int & @Harvard introduce TRAILS — a hidden Markov model that accurately infers time-resolved….
journals.plos.org
Author summary DNA sequences can be compared to reconstruct the evolutionary history of different species. While the ancestral history is usually represented by a single phylogenetic tree, speciation...
0
5
0
And if you are interested in using phase-type distributions, we have implemented a flexible and user-friendly R package called PhaseTypeR (17/18)
joss.theoj.org
Rivas-González et al., (2023). PhaseTypeR: an R package for phase-type distributions in population genetics. Journal of Open Source Software, 8(82), 5054, https://doi.org/10.21105/joss.05054
1
0
2