Jason Chin
@infoecho
Followers
4K
Following
2K
Media
785
Statuses
17K
Common thing in the following subjects: physics, computation, bioinformatics, systems bio., software, engineering? -- A curious mind. The opinions are my own.
San Francisco Bay Area, CA
Joined April 2009
A group of us have been thinking about how to explore human pangenome scale data and make them easier to understand for while. Great collaboration with @GenomeInABottle @sedlazeck @AsifKhalak @srbehera11.
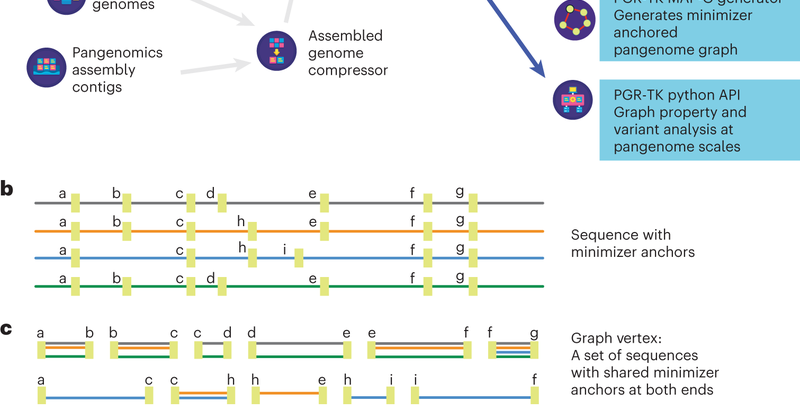
The PanGenome Research Tool Kit (PGR-TK) achieves flexible and scalable representation, visualization, and analysis of genomic variation using pangenome graphs. @infoecho @sedlazeck @GenomeInABottle @psudmant @srbehera11 .
6
33
98
.@headinthebox wondering your thought about this.
julsimon.medium.com
Fool me once, shame on you; fool me twice, shame on me.
0
0
1
PGR-TK code: PGR-TK paper: (with a secret working title: "A renormalization group analysis and toolkit for pangenomes". 🤣).
nature.com
Nature Methods - The PanGenome Research Tool Kit (PGR-TK) achieves flexible and scalable representation, visualization and analysis of genomic variation using pangenome graphs.
1
0
2
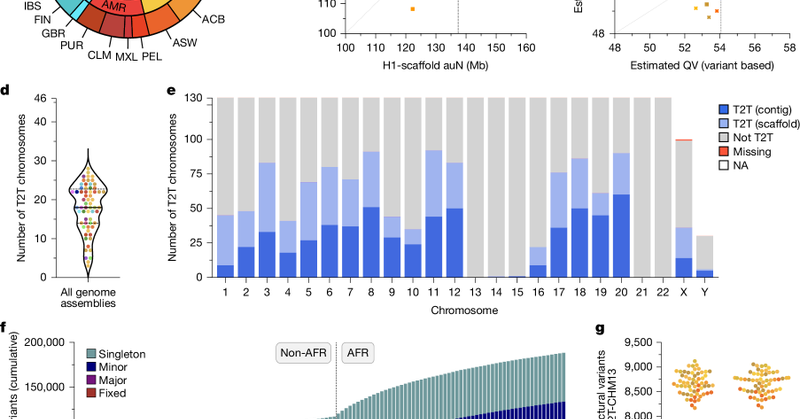
Ref. Thank the HGSVC for me to participate even though I did not hold any affiliation with any research funding.
nature.com
Nature - Using sequencing and haplotype-resolved assembly of 65 diverse human genomes, complex regions including the major histocompatibility complex and centromeres are analysed.
1
1
1
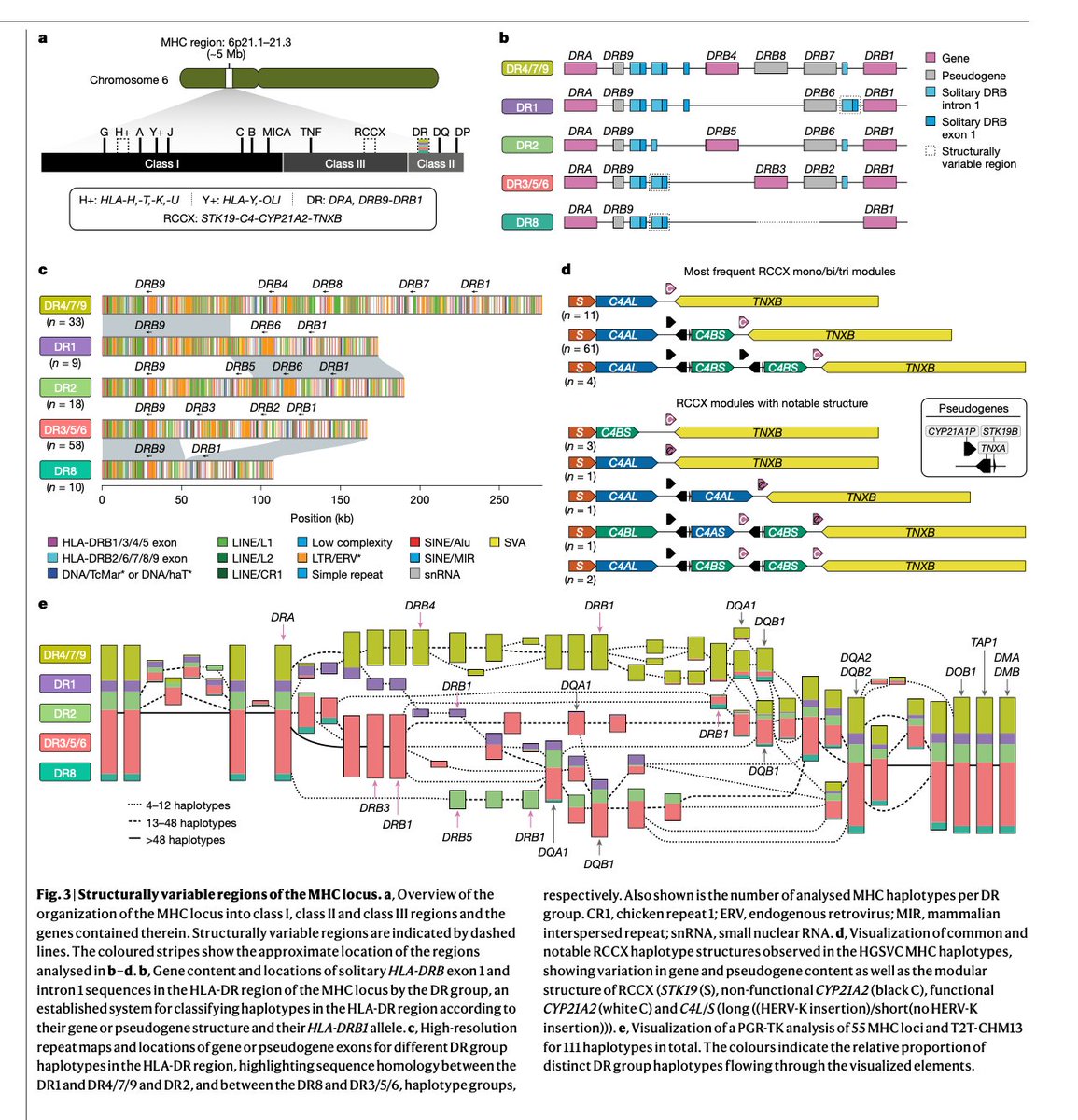
One of the most complex regions in the human genome revealed. To me, it was fascinating to combine genome assembly and pangenome graphs (PGR-TK in Fig. 3e) algorithm techniques to uncover the hidden diversity across human populations. #genomics #bioinformatics
2
42
188
From “modern” ML/AI point of view, is this just a complicated loss function that Nature (maybe a computers from a supreme being) knows to how to optimize?.
→ The Universe Explained. Your life in a nutshell. Particle physics in one equation. Credit: I didn’t upload this image to 𝕏. I’m pointing to an image residing in the 𝕏 stream of “BrianRoemmele” (Apr 14, 2022).
0
0
0