Hunter Bennett
@hbennett_mdphd
Followers
120
Following
140
Media
5
Statuses
30
Intern @StanfordMedRes & Stanford TIP MD/PhD with @UCSDGlassLab and @UCSDSOM Interested in Transcription, Innate Immunity, & Oncology Surfer of waist-high waves
San Diego
Joined January 2023
RT @FunchainMD: Welcome to the incoming @StanfordCancer hematology/oncology fellows! We are excited to keep so many stellar physician scie….
0
5
0
RT @StanfordMedRes: Last batch from the kickoff! We’re excited to meet all of our applicants this season! 🤩🤩 (3/3)
0
2
0
Despite the blowout and the death of the PAC12, we had a blast hosting a @StanfordMedRes tailgate last weekend!
0
1
14
RT @JoyYWu: Welcome to our new @StanfordMedRes interns on their 1st day of residency! And congrats to the new R2s, R3s & chief residents.….
0
8
0
it was a pleasure being involved with this amazing project from @CeeZeeH and @UCSDGlassLab!.
1
0
8
RT @TyTroutman3: My mentor/colleague @UCSDGlassLab and I put together a research briefing on our recent article in collaboration with Laur….
0
2
0
RT @TyTroutman3: I am happy to share my newest publication is online today in the journal @NatImmunol. This was a joint effort with my coll….
nature.com
Nature Immunology - The mechanisms by which noncoding genetic variation shapes tissue macrophage phenotypic diversity remain obscure. Glass and colleagues define cell-intrinsic and environmental...
0
2
0
But all in all so grateful to have been involved in this work, and thankful for the help and expertise of my co-authors @TyTroutman3 and @Enchen__Zhou, and my PhD mentor @UCSDGlassLab. Congrats to all!.
0
0
0
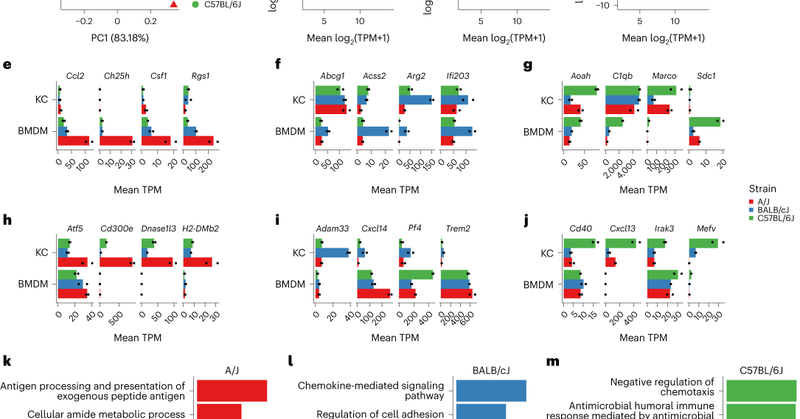
So happy to finally have my thesis work with the world. Our team asked how naturally occurring genetic variants in inbred strains of mice effect transcription and epigenetic regulation in vivo, using Kupffer cells as a model system
nature.com
Nature Immunology - The mechanisms by which noncoding genetic variation shapes tissue macrophage phenotypic diversity remain obscure. Glass and colleagues define cell-intrinsic and environmental...
4
5
26
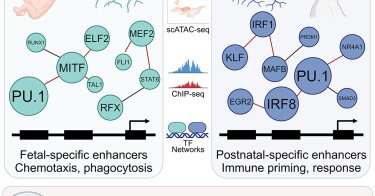
Lead authors @CeeZeeH and Rick Li used organoids, human microglia, and xenotransplantation to detail how microglial networks change from embryogenesis through adulthood. I had a small part of the overall paper, but so thrilled at how it turned out.
1
0
2
Years ago I began to use NicheNet from @saeyslab to analyze some transcriptional data from .@UCSDGlassLab. One of the most interesting uses was the microglial niche. It's great to see that analysis as part of this amazing paper from @CoufalNicole and @UCSDGlassLab in Immunity. .
1
0
3
RT @Pluchinolab: Human microglia maturation is underpinned by specific gene regulatory networks: Immunity
cell.com
Molecular mechanisms that drive human microglia diversity remain largely unknown. Han and Li et al. characterize transitions in the transcriptomes and epigenomes of human fetal and postnatal microg...
0
5
0
RT @gchertow: With spanking new brilliant interns @davidjhwu @hbennett_mdphd and their beloved better halves enjoying BBQ on call. @Stanfor….
0
3
0
RT @StanfordMedRes: Our 2023 Stanford Internal Medicine Residency Interns are starting soon! To celebrate, we reminisced on the moment our….
0
5
0