Guolin Ke
@guolin_ke
Followers
1K
Following
386
Media
27
Statuses
111
@TheDPTechnology, AI for Science. Created #LightGBM, #Graphormer, #UniFold, Uni-Mol, Uni-3DAR. Former Senior Researcher @MSFTResearch. Opinions are my own.
Beijing
Joined February 2017
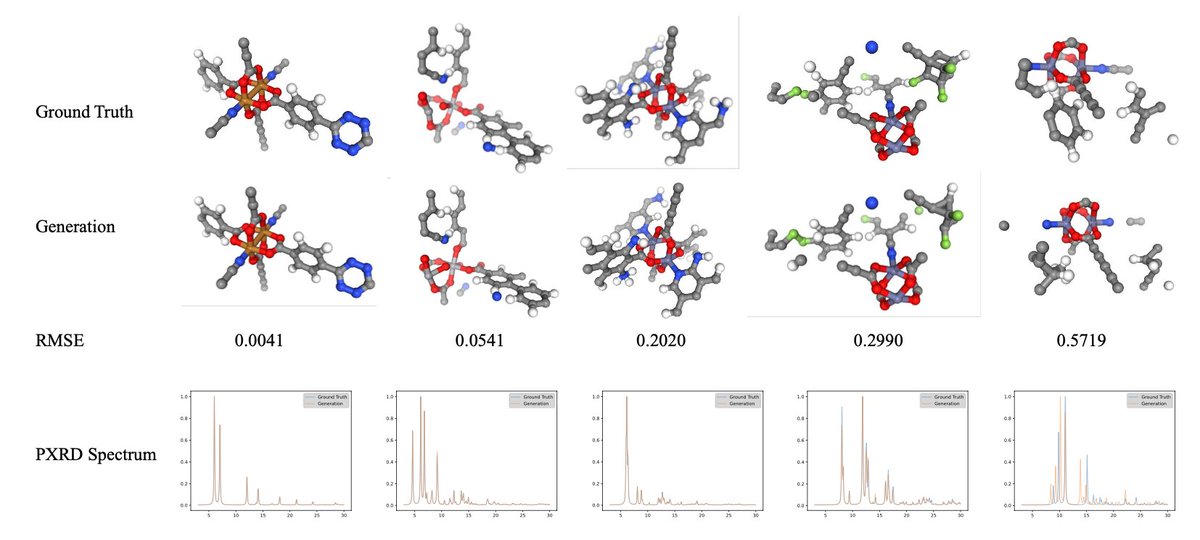
The world is 3D—and many scientific laws are rooted in 3D space. If we want a world model, 3D structure modeling is essential. Yet today, there’s no universal 3D representation—not across scales, not even within microscopic systems. We present Uni-3DAR, a unified 3D framework
0
2
16
RT @wangleiphy: Autoregressive model: alphabets, actions, and atoms This lecture tried to offer a unified perspecti….
0
8
0
The model, training scripts, and dataset for small molecule generation (QM9) with Uni-3DAR are now released!.Check it out on GitHub:
github.com
Implementation of paper "Uni-3DAR: Unified 3D Generation and Understanding via Autoregression on Compressed Spatial Tokens" - dptech-corp/Uni-3DAR
The world is 3D—and many scientific laws are rooted in 3D space. If we want a world model, 3D structure modeling is essential. Yet today, there’s no universal 3D representation—not across scales, not even within microscopic systems. We present Uni-3DAR, a unified 3D framework
0
0
6
RT @BiologyAIDaily: Beyond Atoms: Enhancing Molecular Pretrained Representations with 3D Space Modeling. - Molecular pretrained representat….
0
5
0
RT @Bohrium_AI4S: Transform your #DrugDiscovery with VD-Gen! A cutting-edge tool generates high-affinity 3D molecules using virtual dynamic….
0
4
0
RT @rkakamilan: [2405.11769] Uni-Mol Docking V2: Towards Realistic and Accurate Binding Pose Prediction.みんな大好きUniシリーズのUniMol アプデ.PoseBuster….
0
9
0
Excited to share XtalNet, an approach to predict/generate crystal structures end-to-end from PXRD data, without the need for external databases or extensive manual effort. Check for more details.
arxiv.org
Powder X-ray diffraction (PXRD) is a prevalent technique in materials characterization. While the analysis of PXRD often requires extensive human manual intervention, and most automated method...
Introducing #XtalNet by @guolin_ke and team: E2E crystal structure prediction from PXRD data with contrastive learning and #diffusion based conditional crystal structure generation. paper: dataset:#AI4Sci #GenAI #Crystallography
1
2
27
Thanks @_akhaliq for highlighting our work! 🌟 Uni-SMART tackles analyzing papers with multimodal elements, like molecular graphs & charts, a challenge for LLM. Learn more: .
Uni-SMART. Universal Science Multimodal Analysis and Research Transformer. In scientific research and its application, scientific literature analysis is crucial as it allows researchers to build on the work of others. However, the fast growth of scientific knowledge has led
0
1
17
RT @tri_dao: Announcing FlashAttention-2! We released FlashAttention a year ago, making attn 2-4 faster and is now widely used in most LLM….
0
662
0
RT @momijiame: LightGBM v4 の CUDA 実装、めちゃ速い。HIGGS データセットを 1,000 イテレーション回すのにかかる時間が CPU (Core i7-12700) で 422 秒なのに対し GPU (RTX3060) で 79 秒になった。….
0
26
0
🚀Uni-Mol+ has been updated (, now it is the best model on both PCQM4MV2 & Open Catalyst 2020 IS2RE Direct benchmarks, surpassing previous models by a large margin!. Model & code released (, making it a perfect choice for #NeurIPS
2
4
28
Excited to introduce a new member to the "Uni-" family: Uni-RNA (! It's a large model pre-trained on 1-billion RNA sequences. Uni-RNA achieves state-of-the-art performance in structural, functional, and mRNA-related tasks. Model & applications coming.
biorxiv.org
RNA molecules play a crucial role as intermediaries in diverse biological processes. Attaining a profound understanding of their function can substantially enhance our comprehension of life’s...
3
12
56
Previous Twitter for Uni-Dock.
Uni-GBSA: an open-source and web-based automatic workflow to perform MM/GB(PB)SA calculations for virtual screening. Check it out at Briefings in Bioinformatics @OxfordJournals . #compchem #drugdiscovery #mmgbsa .
0
0
1