Sergei Grudinin
@grudinin
Followers
241
Following
95
Media
3
Statuses
34
Structural bioinformatician, group leader @inria_grenoble and @CnrsAlpes
Grenoble
Joined July 2009
BAnG: Bidirectional Anchored Generation for Conditional RNA Design 1/ This study introduces RNA-BAnG, a deep learning model for designing RNA sequences that interact with specific proteins without requiring extensive experimental data or RNA structural information. 2/ The
1
4
7
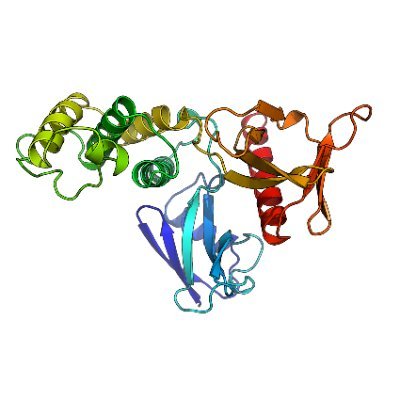
We engineered membrane protein bacteriorhodopsin to be soluble. It binds retinal and can photocycle! X-ray structure reveals conserved binding pocket with 0.8 Å all atom RMSD to WT BR. All this became possible due to hard work by Andrey Nikolaev and the team 🙏 #proteindesign
3
23
122
Come to my group!!! #PhDstudent wanted in an excellence research center in beautiful Barcelona!!!!
Want to do a doctorate at @icmabCSIC ? This is a great opportunity.
0
2
10
Protein sequences to motions, great PhD work by Valentin Lombard, with @grudinin! We investigated whether compact continuous representations of protein motions can be predicted directly from sequences, without relying on or sampling protein structures. https://t.co/OJ93WKacGm 1/3
biorxiv.org
How protein move and deform determines their interactions with the environment and is thus of utmost importance for cellular functioning. Following the revolution in single protein 3D structure...
1
20
106
Very happy to expand our Deep Local Analysis series! Protein interfaces represented as many residue-centred locally oriented cubes. Step 1: what's in the cube? Step 2: what's the effect of mutating that residue? thanks to @yassermhbh @acarbone16 for the great collaboration!
A simple 3D #DeepLearning model with cool ingredients, #DeepLocalAnalysis, to estimate effects of #mutations on #protein-protein interactions https://t.co/zfaV0Aj7kM great collaboration with @yassermhbh @LaineElodie at @LCQB_UMR7238
0
4
30
PhD proposal to work on alternative splicing and protein design. Selected candidates will be funded for 4 years (pre-doc+PhD). Apply before March 24! https://t.co/1OyJiuso5F
https://t.co/8ibcAeb1IB
@iBio_Sorbonne @grudinin
0
3
5
Now we have updated (and submitted9 our AF2 Fold and Dock manuscript. The major change is that we now also benchmark the capacity of AlphaFold2 to separate interacting from non-interacting proteins. https://t.co/hcM7ocDdSm
#Alphafold2
biorxiv.org
Predicting the structure of interacting protein chains is fundamental for understanding the function of proteins. Here, we examine the use of AlphaFold2 (AF2) for predicting the structure of hetero...
0
7
15
Thematic School 2021 : Graph as models in life sciences: #Machinelearning and integrative approaches 📍Online ⏰25th – 29th of October 2021 @ljacob @grudinin @JeanRaisaro @cazencott
1
3
2
What are the ingredients for protein partner identification? Check out our preprint reporting on a molecular cross-docking-based approach for the ab initio reconstruction of protein-protein interaction networks! @acarbone16 @yassermhbh @LCQB_UMR7238
https://t.co/eYWUFoRYEW
1
4
12
Protein language model embeddings for fast, accurate, alignment-free protein structure prediction https://t.co/vPlLggjgFA
#biorxiv_bioinfo
1
8
37
After 2 weeks of working together intensively, we proudly present our assessment of using #alphafold2 from @DeepMind for peptide-protein docking: it works! https://t.co/WAlrvtHccP 🧵
biorxiv.org
Highly accurate protein structure predictions by the recently published deep neural networks such as AlphaFold2 and RoseTTAFold are truly impressive achievements, and will have a tremendous impact...
7
94
372
Proud to share the first work of @zhemchuzhnikovd with assitance from @ilvailvail on our vision of 6DCNNs, roto-translational convolutions, local SE(3) equivariance and a mix of protein representations! https://t.co/TaqndG61ht
#6dcnn #equivariance
1
4
33
by popular demand, @sokrypton is going teach us how to use #RoseTTAFold and #AlphaFold2 on Wednesday, August 4th at 7pm EDT Sergey Ovchinnikov "ColabFold - Making protein folding accessible to all via Google Colab!" https://t.co/b2cWQHIxMr
9
75
252
Our work with @ilvailvail and @nv_pavlichenko is published today! What a day for #AI and protein structure prediction :)
Great new work by @ilvailvail @nv_pavlichenko @grudinin @inria_grenoble @CnrsAlpes @UGrenobleAlpes - 'Spherical convolutions on #moleculargraphs for #protein model quality assessment' - https://t.co/HtTHcRhlsv
#bioinformatics #compchem #imaging #neuralnetworks #deeplearning #AI
1
4
18
Last year we presented #AlphaFold v2 which predicts 3D structures of proteins down to atomic accuracy. Today we’re proud to share the methods in @Nature w/open source code. Excited to see the research this enables. More very soon! https://t.co/6uiV51Xly5
https://t.co/CLo7EKubBT
78
2K
6K
The meeting cost is $200 ($100 students, $1000 industry) plus accommodation charges (€105 per day, including all meals). Accompanying persons are accepted. Paiement details are communicated after pre-registration validation.
0
1
0
The workshop aims at bringing together researchers from computational structural biology and several machine learning communities. As in-person space is limited, participation will have to be approved by the organizers (please answer the two pre-registration form questions).
1
1
5
Structural biology is evolving fast! With the big push coming from machine learning, what are the new challenges? If you’re interested, please consider participating in a small in-person gathering in the south of France this summer!
2
14
55
@MoAlQuraishi @cwcoley Thanks, our perspective (for the casp issue) can be found here
arxiv.org
The potential of deep learning has been recognized in the protein structure prediction community for some time, and became indisputable after CASP13. In CASP14, deep learning has boosted the field...
0
2
7