Edward Chuong
@edchuong
Followers
2K
Following
3K
Media
69
Statuses
648
Asst Prof at CU Boulder studying genomics, transposons, and immunology. UCSD BS - Stanford PhD - Utah Postdoc. He/him. @edchuong.bsky.social
Boulder, CO
Joined February 2013
I’m excited to share our latest study led by @GiuliaPasquesi out today in @CellPressNews , uncovering a new way transposons have been repurposed for human interferon signaling! Read on for a thread on cryptic splice variants, decoy receptors, and viruses (1/N) 👇🧵
13
93
434
Start the new year with a new publication! Led by the talented postdoc @YoseopYoon ! RBBP6 anchors pre-mRNA 3′ end processing to nuclear speckles for efficient gene expression: Molecular Cell
6
11
77
Our new paper defining the role of H3K36 methylation in regulating cell fate and plasticity is available online: https://t.co/WOp1OOH8qi Thanks to @ar_swearingen and our entire team for their tireless work and to the editors at @NatureCellBio for the opportunity!
1
5
19
Check out our latest pre-print: we show that the LINE-1 retrotransposon is a potent source of chromosomal rearrangements and instability in addition to insertional mutagenesis using shotgun and long-read WGS. @KathleenHBurns @JenniferAKarlow
https://t.co/Bu5cod9vbc
biorxiv.org
LINE-1 (L1) retrotransposition is widespread in many cancers, especially those with a high burden of chromosomal rearrangements. However, whether and to what degree L1 activity directly impacts...
6
39
143
See also a highly complementary deep dive into exonization in the same issue of Cell by @YagoArribas
0
2
14
All credit for this discovery goes to amazing postdoc @GiuliaPasquesi , who noticed the isoform in her 2020 Lockdown SideProject (tm) and drove this project start to finish. It was an honor to be a part of it. Look out for her--she's on the job market! 😉
1
2
18
Beyond IFNAR2, our findings suggest that TE exonization may be a widespread yet hidden source of decoy isoforms that regulate immune signaling.
1
0
4
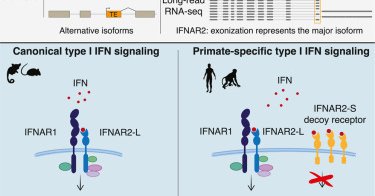
Our study shows how TE exonization gave rise to a primate-specific IFN decoy receptor, which acts as a dial to turn down IFN signaling in human cells. While IFN decoy receptors have evolved repeatedly in viruses (eg VACV B18), this is the first host-encoded IFN decoy receptor.
1
0
4
We also found that risk variants linked to severe COVID-19 in the IFNAR2 locus perfectly coincided with splicing QTLs associated with higher relative expression of the decoy. This suggests that variation in IFNAR2 splicing partly underlies variation in response to infection.
1
0
2
Giulia tested this by using CRISPR to dissect out each isoform, and did a battery of IFN assays. Whenever the short isoform was removed, cells showed more potent IFN responses, including immune gene activation, cytotoxicity, and antiviral effect against DENV-2 & SARS-CoV-2!
1
0
4
Giulia was surprised to see that the short isoform was ubiquitously expressed, often at >2x levels of the canonical isoform! Expression was conserved in primates, and the protein was also detectable by mass-spec, suggesting that this isoform evolved an important immune function.
1
0
5
One caught her eye: a short isoform of the IFN receptor IFNAR2 formed by an exonized Alu. This isoform was first discovered over 20 yrs ago (Pfeffer et al 1997), yet has since been forgotten in the literature, presumably assumed to be silent in human cells.
1
0
4
Giulia revisited this assumption, digging into long-read cDNA-seq looking for exonized TEs that form isoforms with robust expression (>5 TPM) and mass-spec evidence. She uncovered hundreds of isoforms of protein-coding immune genes, which were mostly novel or poorly characterized
1
0
6
Most introns are littered with TEs, which can become “exonized” and create cryptic isoform variants. Yet while thousands of exonization events have been detected, only a handful have been studied in detail, with most variants assumed to deleterious or nonfunctional.
1
1
7
Enter transposable elements (TEs) -- the parasites of the genome! While most TEs are non-coding, they are now appreciated as a source of species-specific regulatory elements, which can drive differences in immune gene regulation. But what about TEs in gene exons?
1
0
3
Interferon (IFN) signaling is an innate immune pathway important for fighting viruses and diseases like cancer. Variation in IFN activity is strongly linked to differences in disease severity including severe COVID-19. What are the factors that influence IFN variation?
1
0
4
Paper Link: "Regulation of human interferon signaling by transposon exonization"
cell.com
Transposable element exonization can yield functional protein isoforms as seen for primate-specific IFNAR2.
1
3
16
Big thanks to collaborators @LilyNNguyen @earth2olive @bitlerlabucd and @NIGMS and @AmericanCancer for supporting this work!
1
0
1
Interestingly, we also found that the key regulatory sequences within LTR10s are contained within an unstable VNTR, which shows both somatic and germline variation, potentially underlying differences in their regulatory activity.
1
0
0