Nick Irwin
@dinokaryon
Followers
545
Following
608
Media
38
Statuses
134
Junior Group Leader @gmivienna @viennabiocenter eukaryotic evolution | horizontal gene transfer | cell biology | plant biology | phylogenetics
Joined November 2017
RT @Luisjagago: Our study on Diphyllatea (CRuMs) is out! From our lovely trip to the South Pacific, we manage to get 4 new transcriptomes f….
0
6
0
RT @delaconcepcionj: How can protein complexes diversify without compromising their function? In our new pre-print, we explore this questio….
biorxiv.org
The evolution of cellular complexity hinges on the capacity of multimeric protein complexes to diversify without compromising their ancestral functions. A central question is how individual subunits...
0
32
0
RT @PlantoPhagy: 🆕 & 🆒#Preprint led by @delaconcepcionj adressing a key #evocellbio question: How can individual subunits of multimeric pro….
0
88
0
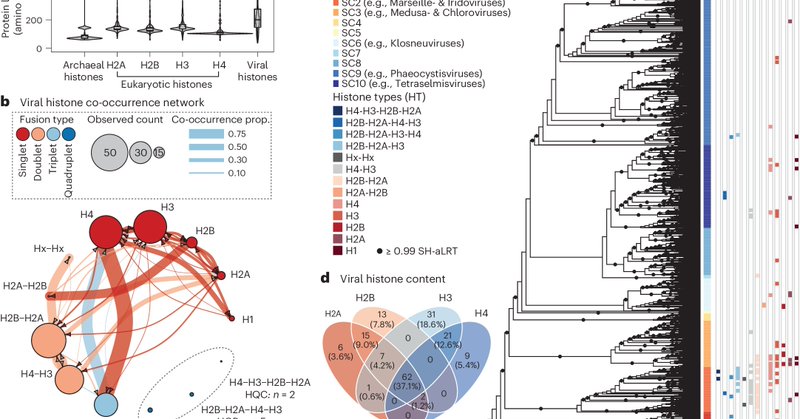
Happy to see our paper on viral histones and the origin of the nucleosome finally out @NatureMicrobiol!. Many thanks to Tom Richards and @MertonCollege.
nature.com
Nature Microbiology - Phylogenetic and functional analyses suggest that viral histones were acquired from early eukaryotes during eukaryogenesis and that nucleosome evolution proceeded through...
Excited to share our manuscript about how we studied the function and evolution of giant viral histones to understand the origin of the nucleosome (1/10)
9
52
159
If you find this work and horizontal gene transfer interesting and want to get involved, get in touch! This will all be continuing next year at my new lab at @gmivienna! (10/10)
1
14
40
Please read the paper for more information! This was a great collaboration with Tom Richards, and many thanks to @tobias_warnecke @antoine_hocher @Luisjagago for help and discussions and @MertonCollege for support! (9/10).
1
0
7
Our phylogenetic analysis of these histone repeats corroborates work by @PaulTalbert10 and @AlbertErives showing that these viral histones branch between Archaea and eukaryotes in an intermediate position suggesting an ancient origin (3/10)
2
1
10
RT @gmivienna: We are #hiring! The Gregor Mendel Institute has a Junior Group Leader position available. We seek #researchers who will help….
0
79
0
So excited to be joining the GMI next year! 🫛.
🎉 Exciting news! New group leaders will join the GMI next year. Today we introduce Nick Irwin (@dinokaryon), whose lab will investigate horizontal gene transfer in plants and algae. 🌱 Nick’s group page:
9
6
68
RT @PlantoPhagy: Chuffed to finally have our preprint (Shuffled ATG8 interacting motifs form an ancestral bridge between #UFMylation and #a….
0
60
0
RT @gutmicrobiome: Our paper made the cover! . One of our beautiful Bryozoa colonies from Quadra Island in British Columbia collected by @d….
0
11
0
RT @gutmicrobiome: So pleased to see our paper out today in @NatureMicro! .We compared bacterial microbiomes of microscopic marine inverteb….
0
21
0
RT @PlantoPhagy: Here it is🥰 Our new #preprint led by L. Picchianti, @Victor_SMH, N. Zhan together with @gekaragoz lab (the first of many t….
0
62
0