Ogün Adebali
@compGenomics
Followers
2K
Following
2K
Media
179
Statuses
945
Associate Professor at Sabancı University
Istanbul, Turkey
Joined January 2016
1/n UV radiation disrupts genome integrity, but how does the 3D genome facilitate DNA damage response (DDR)? Our latest study with @vogulcankaya reveals how UV-induced genome reorganization enables efficient repair. 🌟🔬 🔗 https://t.co/xDcz9dX1P1
3
18
85
Have you already read this review? Conserved 3D genome reorganization during DNA repair @compGenomics @uzh_vetsuisse @fenssabanci @vogulcankaya
https://t.co/aotuvV4f6x
0
1
3
Just realized that our work was highlighted - some time ago - by a popular science platform. Written by a real person, not AI 🥰 https://t.co/BqHvRTgSIR
azolifesciences.com
Research shows UV radiation modifies 3D genome structure, affecting DNA damage response and gene activity, offering insights for cancer prevention strategies.
0
0
4
I’m happy to co-author this review with our PhD candidate, @vogulcankaya! A big thanks to Drs Nägeli and Yancoskie for their lead. We’ll be sharing more updates from this team in the coming months. Stay tuned!
This review synthesizes findings from genomic, molecular, and disease studies that chromatin organization dynamically shapes diverse DNA repair pathways. @compGenomics @uzh_vetsuisse @fenssabanci @vogulcankaya
https://t.co/d6Yy9XNWzG
0
2
11
Brendo: an open resource for uniquely transcribed genes in brain endothelial cells https://t.co/ZgfGl1QqOK
#biorxiv_bioinfo
biorxiv.org
The blood-brain barrier (BBB) plays a critical role in central nervous system homeostasis, yet comprehensive transcriptional profiling of brain microvascular endothelial cells (BMECs) remains...
0
3
2
(2/n) Oct 20 at @itu1773 Öncelikli ve Yenilikçi Araştırmalar Çalıştayı Nov 19 at @BilkentUniv MBG Departmental Seminar Nov 21 at @METU_ODTU Departmental Seminar There might be a couple more, currently uncertain... Feel free to join.
0
0
4
After a year away on my sabbatical, I accepted every seminar invitation that I could. Here is the schedule: (1/n) Oct 4 at @itu1773 BioScenario Oct 14 at Istanbul Univ - Evolution Club Oct 17 at @kocuniversity MBG Departmental Seminar Oct 18 at @UniBogazici BUEC IEEE
1
0
7
Bad news for me. I can’t be rude, as it feels like speaking with a real person.
Rude prompts to LLMs consistently lead to better results than polite ones 🤯 The authors found that very polite and polite tones reduced accuracy, while neutral, rude, and very rude tones improved it. Statistical tests confirmed that the differences were significant, not
1
0
2
In the lab, we began using @RocketChat for internal communication. We are self-hosting. So far, I loved it! We are also self-hosting @getoutline for internal documentation and training purposes. It is elegant, simple, easy to set up and useful.
0
0
11
8/ 🔓 #OpenScience All code, data, and benchmark scripts are freely available: https://t.co/pvRS6lzA66 PHACE predictions:
zenodo.org
The co-evolution trends of amino acids within or between genes offer valuable insights into protein structure and function. Existing tools for uncovering co-evolutionary signals primarily rely on...
0
0
2
7/ Key insight: By eliminating phylogenetic dependency, PHACE uncovers genuine co-evolution, not artifacts of shared evolutionary history. See examples in the paper why PHACE succeeds.
1
0
2
6/ Benchmarking: PHACE is compared to the current methods (like DCA, PSICOV, GaussDCA, MIp, CAPS, CoMap) in identifying co-evolving residues across >650 proteins. demonstrating high accuracy (MCC, AUC, F1) and robustness to alignment size/divergence.
1
0
2
5/ How does PHACE work? 👉 Uses ancestral sequence reconstruction 👉 Traverses the tree to detect true independent amino acid changes 👉 Groups tolerated amino acids to filter out neutral variation 👉 Weights evolutionary branches by their actual rate of change
1
0
2
4/ Why does this matter? Most co-evolution tools ignore shared ancestry, so a single ancestral change can be mistakenly counted as multiple co-evolution events. PHACE tracks real, independent substitutions, avoiding this trap.
1
0
2
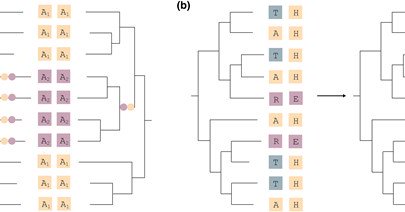
3/ We introduce PHACE, a new algorithm that: ✔️ Maps amino acid substitutions onto phylogenetic trees ✔️ Identifies phylogenetically independent co-evolution events ✔️ Categorizes amino acids as 'tolerable' or 'intolerable' at each site ✔️ Explicitly models gaps as a third state
1
0
2
2/ Co-evolution between amino acid positions offers vital clues about protein structure and function. However, traditional methods often overlook evolutionary relationships, resulting in misleading signals. We aimed to fix this.
1
0
1
I'm excited to announce our new paper: PHACE: Phylogeny-Aware Detection of Molecular Co-Evolution by Nurdan Kuru & me, just published in Molecular Biology and Evolution! https://t.co/yes183VQU9
#Phylogenetics #CoEvolution #Proteins #Bioinformatics
academic.oup.com
Abstract. The coevolution trends of amino acids within or between genes offer key insights into protein structure and function. Existing tools for uncoveri
1
4
29
In Boston next three days. Giving a talk tomorrow. If you’re around, drop a message.
1
0
38
Nice artwork. Replace "conference" with "journal". I guess the donkey would be the reviewers. Wait... how about the authors who pay insane amounts?
Nature publishing a piece on unpaid academic labor is the best joke I’ve read in a while. https://t.co/hLK5opHafC
0
1
5