Christof Angermüller
@cangermueller
Followers
1K
Following
7
Media
184
Statuses
694
Software Engineer at Google
Mountain View, CA
Joined November 2012
Excited to announce our latest paper on ML-guided design of single-domain antibodies against CoV-2; A collaboration between @GoogleDeepMind, @AAlphaBio and @LumenBio.
1
14
96
RT @DdelAlamo: "Sensitive remote homology search by local alignment of small positional embeddings from protein language models". ESM2 embe….
0
21
0
Contextualizing protein representations using deep learning on protein networks and single-cell data.
biorxiv.org
Understanding protein function and discovering molecular therapies require deciphering the cell types in which proteins act as well as the interactions between proteins. However, modeling protein...
0
2
7
The Patent and Literature Antibody Database (PLAbDab): an evolving reference set of functionally diverse, literature-annotated antibody sequences and structures.
biorxiv.org
Antibodies are key proteins of the adaptive immune system, and there exists a large body of academic literature and patents dedicated to their study and concomitant conversion into therapeutics,...
0
1
8
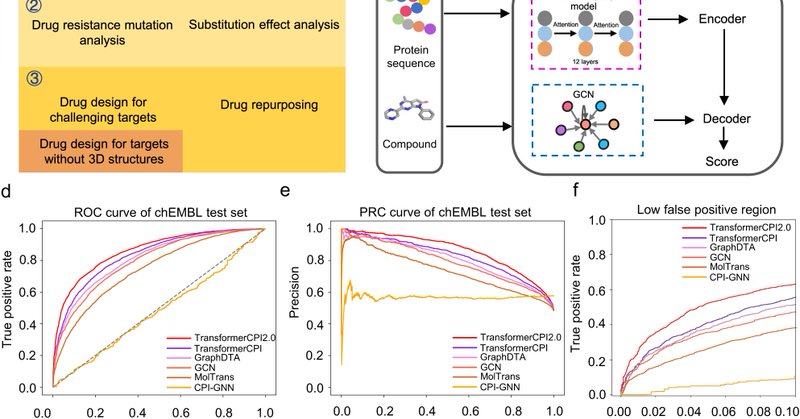
Sequence-based drug design as a concept in computational drug design.
nature.com
Nature Communications - Conventional structure-based drug design pipeline is a complex, human-engineered process with multiple independently optimized steps. Here, the authors report a...
0
19
75
RT @DdelAlamo: “Large language models are universal biomedical simulators”. Another use of GPT-4 for biomedical sciences research. https://….
0
25
0
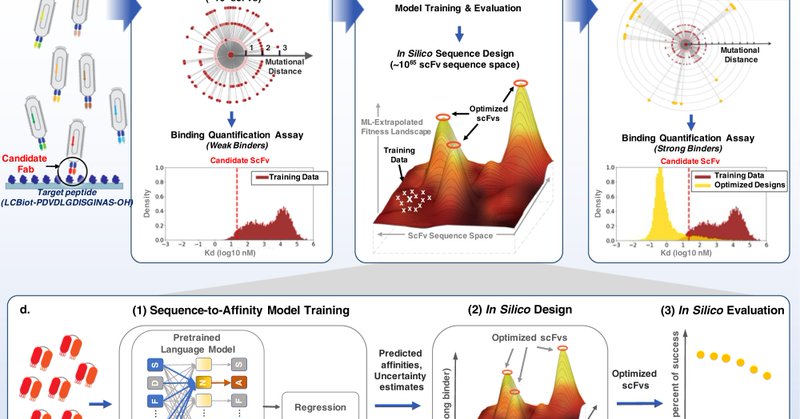
Machine learning optimization of candidate antibody yields highly diverse sub-nanomolar affinity antibody libraries.
nature.com
Nature Communications - Therapeutic antibody discovery is time and cost-intensive. Here, the authors develop a machine learning-driven method enabling accelerated design of large and diverse...
0
4
40
Mol-Instructions: A Large-Scale Biomolecular Instruction Dataset for Large Language Models.
arxiv.org
Large Language Models (LLMs), with their remarkable task-handling capabilities and innovative outputs, have catalyzed significant advancements across a spectrum of fields. However, their...
0
0
7
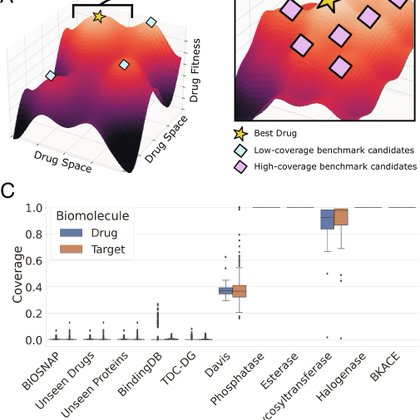
Contrastive learning in protein language space predicts interactions between drugs and protein targets.
pnas.org
Sequence-based prediction of drug–target interactions has the potential to accelerate drug discovery by complementing experimental screens. Such co...
0
0
24
RT @omarsar0: BiomedGPT, a unified biomedical generative pretrained transformer model for vision, language, and multimodal tasks. Achieves….
0
212
0
RT @DdelAlamo: “Self-driving laboratories to autonomously navigate the protein fitness landscape”. Four independent trajectories “quickly c….
0
58
0
A general model to predict small molecule substrates of enzymes based on machine and deep learning.
nature.com
Nature Communications - For many enzymes, it is unknown which primary and/or secondary reactions they catalyze. Here, the authors use machine and deep learning to develop a general model for the...
0
10
41