caflischgroup
@caflischgroup
Followers
787
Following
100
Media
19
Statuses
88
Computational structural biology @uzh_ch drug discovery, simulation & advanced sampling, analysis tools devpmt, amyloid research Account managed by group member
Zurich, Switzerland
Joined August 2017
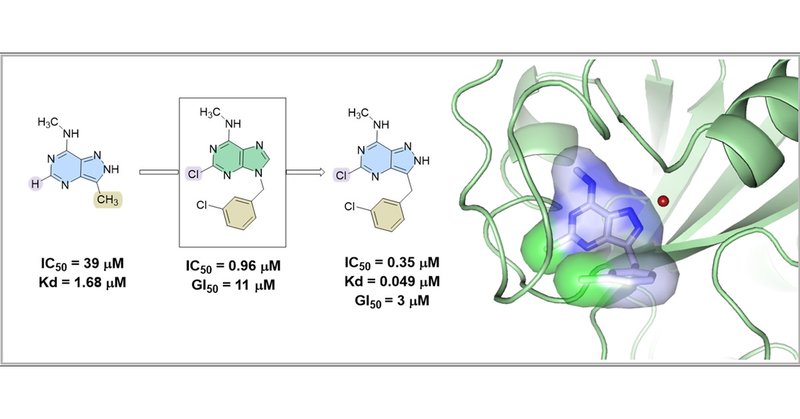
✨🧬Thrilled to announce our latest study on #JMedChem:. A structure-based design of a powerful inhibitor targeting YTHDC1, a key m6A reader. Congrats to @z_frantisek and all the team! #DrugDiscovery . Discover more in the link below 👇👇👇 .
pubs.acs.org
N6-Adenosine methylation (m6A) is a prevalent post-transcriptional modification of mRNA, with YTHDC1 being the reader protein responsible for recognizing this modification in the cell nucleus. Here,...
0
1
1
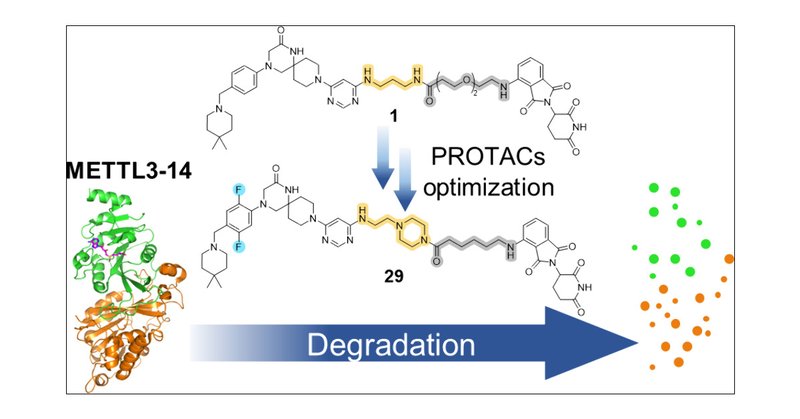
🔬Unveiling the first CRBN-based PROTACs for the METTL3-14 m6A-RNA writer 🧬Opening new possibilities in epitranscriptomics research 🌐.Check out our most recent publication in JACS Au 👇@uzh @JACS_Au .
pubs.acs.org
Methylation of adenine N6 (m6A) is the most frequent RNA modification. On mRNA, it is catalyzed by the METTL3–14 heterodimer complex, which plays a key role in acute myeloid leukemia (AML) and other...
1
4
17
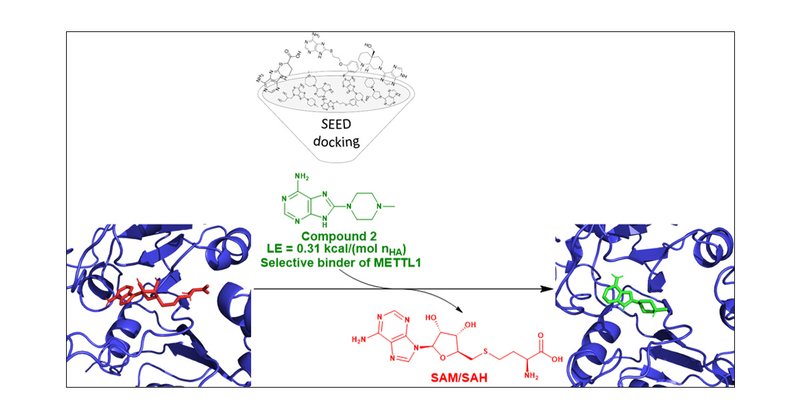
New story out: we reveal the first inhibitors of the m7G-RNA writer METTL1. @ACSBioMed.
pubs.acs.org
We discovered the first inhibitors of the m7G-RNA writer METTL1 by high-throughput docking and an enzymatic assay based on luminescence. Eleven compounds, which belong to three different chemotypes,...
0
2
5
Check out this new work on the reevaluation of bromodomain ligands targeting BAZ2A 👇👇. @NevadoLab @GrazianoLolli @UZH_en @UniTrento .
onlinelibrary.wiley.com
BAZ2A promotes migration and invasion in prostate cancer. Two chemical probes, the specific BAZ2-ICR, and the BAZ2/BRD9 cross-reactive GSK2801, interfere with the recognition of acetylated lysines...
0
0
3
RT @ACSBioMed: Disrupting dimeric β-amyloid by electric fields. By Amedeo Caflisch et al @UZH_en @caflischgroup #Alzheimers. 🔓 Open access….
0
2
0
RT @JPhysChem: Disrupting dimeric β-amyloid by electric fields. By Amedeo Caflisch et al @UZH_en @caflischgroup #Alzheimers. 🔓 Open access….
0
6
0
Please check out the UZH Award for Research in Brain Diseases that will be presented for the 18th time on Thursday, July 13, 2023. All the information down here 👇:.@UZH_en.
de.linkedin.com
Erkrankungen des Gehirns gehören noch immer zu den grossen Herausforderungen der Medizin. Auch dieses Jahr zeichnet die Universität Zürich | University of Zurich drei Nachwuchsforschende für ihre...
0
0
4
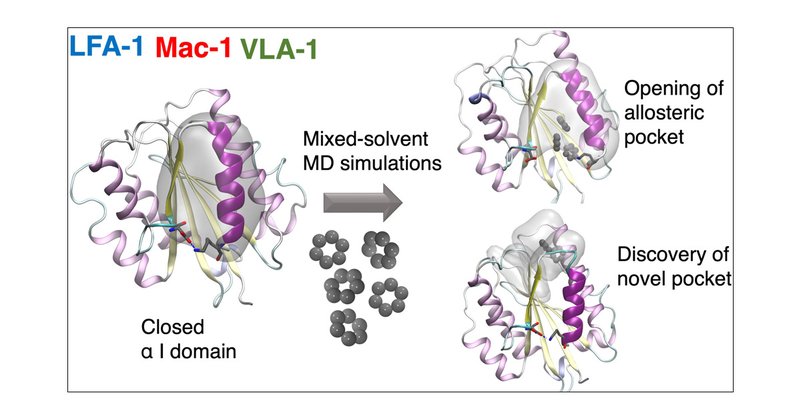
Check our new work on Decrypting Integrins by Mixed-Solvent Molecular Dynamics Simulations @ioana_ilie_UvA @JCIM_JCTC .
pubs.acs.org
Integrins are a family of α/β heterodimeric cell surface adhesion receptors which are capable of transmitting signals bidirectionally across membranes. They are known for their therapeutic potential...
0
5
32
RT @ACSBioMed: Structure-based design of inhibitors of the m6A-RNA writer enzyme #METTL3. By Amedeo Caflisch et al @UZH_ch @caflischgroup ….
0
4
0
Which features of the spiking activity capture better the working memory processes of the human brain? .Check this out in our latest work !.
academic.oup.com
Abstract. Persistent activity has commonly been considered to be a hallmark of working memory (WM). Recent evidence indicates that neuronal discharges in t
0
1
4
We have identified and characterized by x-ray crystallography the first small-molecule ligands of the m6A-reader YTHDF2. Two of them features good ligand efficiency, providing a starting point for hit optimization. @ACSMedChemLett @ACSPublications.
pubs.acs.org
We report 17 small-molecule ligands that compete with N6-methyladenosine (m6A) for binding to the m6A-reader domain of YTHDF2 (YT521-B homology domain family 2). We determined their binding mode at...
0
0
5
RT @ioana_ilie_UvA: Antibody binding makes the prion protein more flexible! #compchem #prions @BBAjournals
https://….
0
2
0
Here, we investigate the influence of the POM1 and POM6 antibodies on the flexibility of the mouse prion protein (PrP) by molecular dynamics simulations. #prions #compchem @ioana_ilie_UvA @BBAjournals .
0
3
9
Molecular dynamics simulations at different temperatures suggest structural reasons for the distinct behavior of two microbial transglutaminases. @clangi3 @amedeocaflisch @CSB_Journal .
0
2
2
Little is known about the potential interactions between flexible tail, globular domain, and the membrane. Here, we used atomistic simulations to investigate how these interactions are modulated. @BiophysJ @ioana_ilie_UvA @MarcoBacci07 @amedeocaflisch .
0
0
9
We used in-silico methods to design binders for the m6A reader protein YTHDC1, leading to highly efficient lead compound and thus pushing the field to the forefront. @ELSchemistry @amedeocaflisch .
0
0
8
Domino effect in allosteric signaling of peptide binding: We apply a novel method to analyse complex MD data and elucidate allosteric pathways without previous knowledge of the system. Check @JMolBiol for more. @pavaro2906 @amedeocaflisch .
0
2
8