busrasavas
@busrasavas
Followers
176
Following
941
Media
12
Statuses
156
PhD student IBG & DEU | Computational Structural Biologist
Izmir, Turkey
Joined February 2020
It's one of those great moments when hard work pays off. Sleepless nights with the most self-motivated team ever!!💪💜 Waiting in huge excitement to be involved the next round of CAPRI, because once you become a CAPRI'ot and taste that adrenaline... 🫠🚀
Super proud to have been listed as one of the successful groups in predicting the structure of an antibody-peptide during @CAPRIdock 's 55th round! This was our first attempt in CAPRI as the Karaca team with @busrasavas @iremylmazbilek @atakanzsn 💪 Here is what we did ...
1
1
17
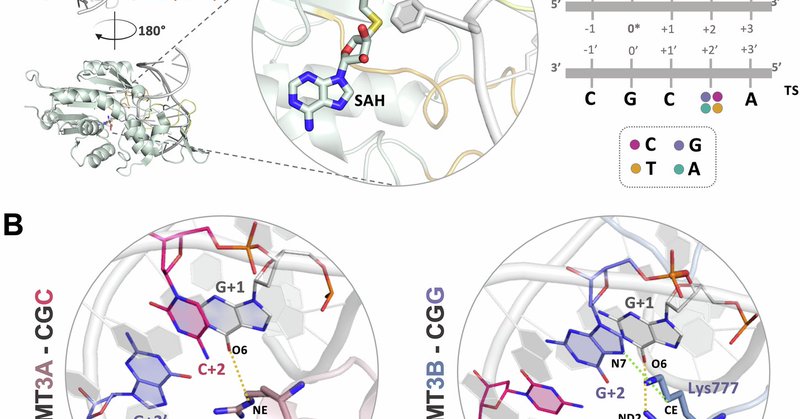
🚀 Excited to share our article with @Ezgi_Karaca_ is now out in @CommsBio! We explored DNA readout rules of almost identical DNMT3A and DNMT3B (91% sequence similarity!), and we asked how can nearly the same proteins “see” DNA so differently? 🧬 https://t.co/jOSh6MGDXp
nature.com
Communications Biology - Comparative dynamics analysis identifies key residues driving sequence specificity differences between human DNA methyltransferases DNMT3A and DNMT3B.
1
2
10
"A stylish woman in a beige coat walking confidently past a moving train at a modern train station." Create images and videos in seconds with Grok Imagine.
831
718
5K
Taken together, our results point to new exciting directions for structural biology through distograms: - Rapid flexibility detection - Improved EM map interpretation - Distogram-informed ensemble modeling Full story 👉 https://t.co/uPTpcO0vDG 💻GitHub:
github.com
Data accompanying our paper entitled "Exploring the Potential of AlphaFold Distograms for Flexibility Assignment in Cryo-EM Maps " - CSB-KaracaLab/af-distogram-flexibility
1
0
1
We next asked: how do AF2-based sampling strategies affect distograms? We tested MinnieFold, AFsample2, and AF_cluster. The biggest impact came from the AF2 version itself. Only AF2.3 mirrored AF3’s flexibility-aware distogram profile.
1
0
2
We run MD of apo and holo AK2 and revealed that holo AK2 hinge distance is sampled rarely in its apo state! Strikingly, apo AF3 distogram reproduces the exact same behavior!!🤯
1
0
1
Surprisingly, AF3 apo and holo AK2 predictions beautifully reflect this conformational change, where C-ter of AK2 experiences a disorder-to-order transition upon beta-stacking itself to AIFM1👇 To track hinge motion, we concentrated on 230-233 CB–CB distances of AK2.
1
0
1
We tested this hypothesis on AK2:AIFM1 complex, a molecular switch between energy metabolism and cell death. 🔁💀 Its function depends on a hinge-driven motion, that is: ✔Recently revealed by two cryo-EM maps ✔ Occurs upon binding ❌Not seen in crystal structures
1
0
1
A primer on distograms: for each residue pair, AF outputs distance probability distributions. Earlier, it was shown that the shape of the distogram peaks may inform us on protein flexibility. How? ✔ Unimodal peak → rigid behavior ✔ Bimodal peak → flexible behavior
1
0
2
🚨 Super excited for our new preprint on flexibility cues in AlphaFold! Together with @Ezgi_Karaca_ and @aysebercinb, we found that distograms of AF2.3 and AF3 mirror MD sampling by predicting the extent of a novel conformational change! 🤯 For more👇
Exploring the Potential of AlphaFold Distograms for Flexibility Assignment in Cryo-EM Maps https://t.co/KWRIZyPXF4
#biorxiv_bioinfo
2
4
33
from my limited experience this morning, an external "cat force field" is critical to getting correct dynamics. 😸 Left = without cat Right = with cat
2
1
24
since (AI-generated) protein folding (videos) is solved, we now tackle protein conformational changes
4
25
234
CASP is getting cut by NIH... 😢 (Anyone with extra funds wanna help support perhaps the most important competition of the century?) https://t.co/mOuROZYJLh
science.org
Agency silent on funding renewal for contest that inspired creation of AIs that predicted how proteins would fold
20
133
345
Only a few days left to apply for our EMBO course with a great lineup of speakers and tutors! Please feel free to RT! Course website: https://t.co/AeGGFwhzHM Organizers: myself, @amjjbonvin @arneelof @LindorffLarsen Location: @ibgmerkezi
0
16
22
We want awe. We want discovery. And we want them now! Huge thanks to @ItaiYanai & @MartinJLercher for the most enjoyable lecture on Night Science — a reminder that opennes lie at the heart of discovery!
We had two great 'Night Science' workshops on the creative scientific process in Heidelberg yesterday (at EMBL and at the EMBO PhD Course). Here are the slides if you're interested in running something like this at your place! https://t.co/HIDjK27nm1
@MartinJLercher
1
1
10
Towards a greener AlphaFold2 protocol for antibody-antigen modeling: Insights from CAPRI Round 55 1. This paper presents MinnieFold, a greener and more computationally efficient version of Wallner’s massive AlphaFold2 (AF2) sampling, designed to predict antibody-antigen
2
6
43
Just accepted for publication in Proteins! Congrats to @busrasavas for her first first author paper! On the CAPRI55 set, we show that even with 95% reduced massive-AF2 (Wallner approach) sampling, you can still get good quality antibody-antigen complexes. For more 👇
Towards a greener AlphaFold2 protocol for antibody-antigen modeling: Insights from CAPRI Round 55 1. This paper presents MinnieFold, a greener and more computationally efficient version of Wallner’s massive AlphaFold2 (AF2) sampling, designed to predict antibody-antigen
0
1
29
The legendary EMBO Integrative Modeling Course is back! If you want to be in this picture in the 2025 edition, just go ahead and register at https://t.co/AeGGFwi7xk (28 Sept – 03 Oct) In this edition, we have @LindorffLarsen on board to properly explore biomolecular dynamics 💪
meetings.embo.org
Experimental structural studies of interactions can be costly, low-throughput, and challenging. Therefore, computational methods aiming to model protein complexes are particularly valuable. The accur…
We still feel the motivational energy we got from being with all these great minds in one of the most impressive ancient cities of the world, Ephesus ❤️ Co-organized by @amjjbonvin & @arneelof, this event was possible thanks to the generous support from @EMBO ...
0
14
41
2024 has been the most challenging year of my life due to a serious health challenge in my family. During this difficult time, these incredible people have been my rock. Thank you all for your support and camaraderie. I am proud to work alongside you. This post is for you! ❤️🍀🧿
1
4
54
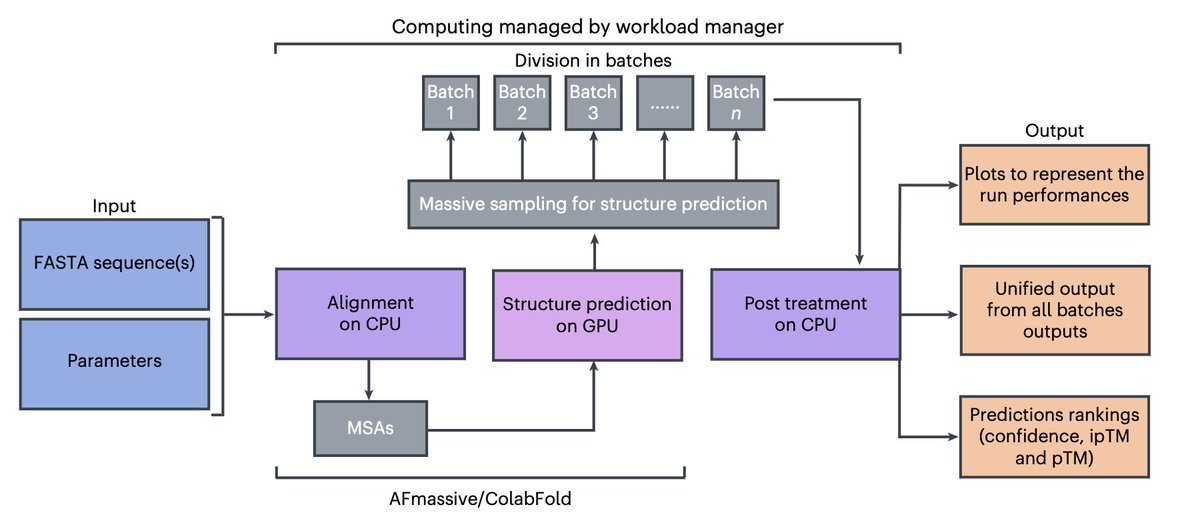
MassiveFold: unveiling AlphaFold’s hidden potential with optimized and parallelized massive sampling @NatComputSci 1. MassiveFold introduces an optimized, parallelized approach to enhance AlphaFold’s structure prediction, significantly reducing computational time for massive
1
13
56
Here we describe our “mini aggressive sampling” approach, which brought us success in CAPRI55’s antibody-peptide target 🎯👇
Towards a greener AlphaFold2 protocol for protein complex modeling: Insights from CAPRI Round 55 https://t.co/hJCYGHpKHX
#biorxiv_bioinfo
1
5
27
Excited to share our latest research on de novo DNA methylation mediated by DNMT3A/B enzymes! @Ezgi_Karaca_ ✨ Check out how a single amino acid difference drives epigenetic selectivity. For more this thread👇&
2
4
23