BMRlab
@bmr_ibr
Followers
282
Following
268
Media
51
Statuses
165
Laboratorio de Biofísica del Reconocimiento Molecular, IBR, Rosario 🧬Biophysics of Molecular Recognition Lab
Rosario, Argentina
Joined March 2022
Welcome to the first of our weekly threads to introduce the Biophysics of Molecular Recognition Lab and our team members. #MeetOurTeam 👇🧵🪡 1/5
1
6
36
Excited to share our latest article published in *Analytica Chimica Acta* @ELSchemistry Multivariate curve resolution-based data fusion approaches applied in ...
4
5
23
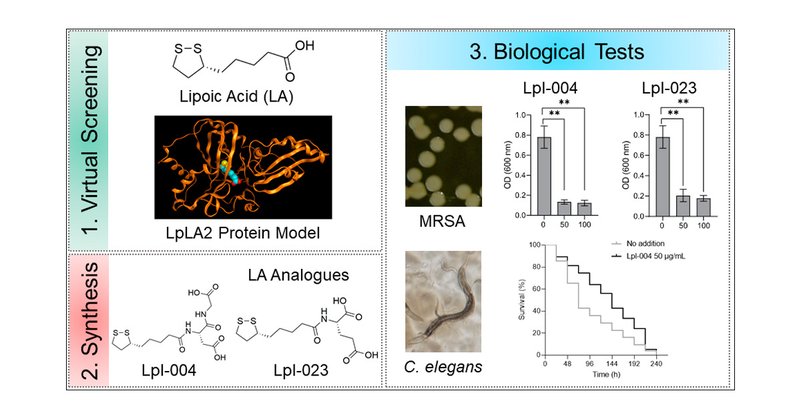
Our contribution from the public university of Argentina @UNRoficial @fbioyfunr and CONICET @CONICETDialoga @IBR_CONICET to the fight against Gram positive pathogens! https://t.co/USLKISZ1LK
pubs.acs.org
Lipoic acid (LA) is an essential cofactor in prokaryotic and eukaryotic organisms, required for the function of several multienzyme complexes such as oxoacid dehydrogenases. Prokaryotes either...
6
8
29
El Dr. Rodolfo Rasia, director del grupo de Biofísica del Reconocimiento Molecular en @IBR_CONICET dialogó con #ABC sobre este trabajo. Los resultados fueron publicados en la revista Biochemistry. La nota complenta en https://t.co/dvP1uOOSKx📲
0
2
10
Muchas gracias @CarlosGBava @341 por difundir nuestro trabajo 😊
UN EQUIPO DEL CONICET DISEÑÓ UNA NUEVA ENZIMA PARA LA INDUSTRIA DEL ACEITE https://t.co/8Zj1kySIqC
@IBR_CONICET
@iprobyq
@FitoDeRosario
0
1
6
👩🔬👨🏽🔬Especialistas de @bmr_ibr @iprobyq y @KeclonSA diseñan una nueva enzima optimizada para la industria del aceite. 🏭Su aplicación permitiría realizar el refinamiento de manera más eficiente y económica. https://t.co/DmLnSRkROr
0
29
65
📢Hoy dimos inicio al ciclo de seminarios IBR 2024! @marchucarletti dió su charla sobre diseño de fosfolipasas y @francobiglione sobre un nuevo método para la clasificación funcional de factores de transcripción en plantas. 👏 @bmr_ibr Más info: https://t.co/NsgAs2Q8lV
0
4
11
It is @francobiglione 's turn with his talk on a new method for functional classification of plant transcription factors
0
3
25
Pau Vacs with her first protein gel and @marchucarletti with her first synthetic genes, working at @bmr_ibr @IBR_CONICET
0
2
37
Thanks to @IBR_CONICET and @iprobyq for hosting the research, @CONICETDialoga @agenciaidiar ASACTeI @gobsantafe for funding, @KeclonSA for productive collaborations, and authors @die_goval89 @LuisinaNardo Fiorella Marchisio, Euge Castelli, @salvapeiru @labriataphd @hugomenzella
0
1
8
Implementation of enzymatic degumming in plants needs an initial capital investments and operational expenses that hinder the widespread implementation of the technique. We showed ChPLC reduces the overall cost of soybean oil enzymatic degumming by 58%
pubs.acs.org
The increasing demand pressures the vegetable oil industry to develop novel refining methods. Degumming with type C phospholipases (PLCs) is a green technology and provides extra oil. However,...
1
0
4
We tried to figure out what makes the consensus enzyme so stable, so we produced several point mutants. None of the mutations changed the melting temperature significantly but all abolished thermal unfolding reversibility
1
0
1
We also found that, in contrast to other homologs tested, consensus PLC thermal unfolding is partly reversible. This also adds to the improved activity in the harsh degumming conditions
1
0
2
In our case the result was impressive. The consensus enzyme is stable up to 95 ºC (18 ºC higher than the most stable natural variant tested), retains full activity both in water and in crude oil, where the activity tops at 80 ºC
1
0
1
An artificial variant with the most probable amino acid at each position in a multiple sequence alignment is expected to yield a protein more stable than any individual protein within the alignment. To put it simply, we played "Wordle" with sequences tested by nature
1
0
1
In a set of sequences with the same fold, neutral or destabilizing mutations have no selective pressure but stabilizing mutations (less probable), are favored
1
0
2
We achieved a substantial thermal stabilization of a PLC by consensus sequence design. The underlying idea of the method, first proposed in 1994, is simple
1
0
1
Oil degumming using type C phospholipases (PLCs) represents an environmentally friendly approach, giving an improved yield. Nevertheless, natural PLCs exhibit limited activity under the rigorous conditions employed in oil refining facilities
1
0
3
Happy to share our latest publication! https://t.co/HckN03AEdS We improved an enzyme that is used in edible oil degumming by consensus sequence design @IBR_CONICET @iprobyq @BiochemistryACS
2
14
43