Bin Zhang
@binzmit
Followers
1K
Following
252
Media
9
Statuses
157
Associate professor of chemistry, MIT
East Cambridge, Cambridge
Joined November 2017
Our paper is now online at Sci Adv: https://t.co/1790lwk9Kh See also the accompanying news: https://t.co/H3EZghJt1V
news.mit.edu
MIT chemists found a new way to determine 3D genome structures, using generative AI, that can predict thousands of genome structures in minutes, making it much speedier than existing methods for...
I am excited to share our latest #preprint: ChromoGen: Diffusion model predicts single-cell chromatin conformations https://t.co/h5FORbb0io As the title suggests, we achieved de novo prediction of 3D chromatin structures using DNA sequence and ATAC-seq using AI.
2
1
31
Hmm, I assume these posts were made by AI? Anyway, very impressive summary of our latest preprint.
Protein Language Model Identifies Disordered, Conserved Motifs Driving Phase Separation 1. This study employs ESM2, a cutting-edge protein language model, to analyze intrinsically disordered regions (IDRs) in proteins, uncovering conserved motifs critical for phase separation
2
0
37
Check out our recent manuscript that studies the interactions between small molecules and biomolecular condensates with all-atom simulations:
pubs.acs.org
Biomolecular condensates are essential in various cellular processes, and their misregulation has been demonstrated to underlie disease. Small molecules that modulate condensate stability and...
0
0
14
Our findings shed light on the significant regulatory roles of bio condensate and DNA linker length and help bridge the gap between in vivo and in vitro observations.
1
2
10
Nucleosome condensate and linker DNA alter chromatin folding pathways and rates [Qiu et al, 2024] https://t.co/2MYkfyk67M ▶️residue-level coarse-grained models; non-Markovian dynamics ▶️10n bp DNA linker lengths favor zigzag fibrils ▶️10n+5 bp chromatin loses unique conformations
1
5
53
Scaling Graph Neural Networks to Large Proteins 1. This paper introduces the DISPEF dataset, specifically designed for benchmarking graph neural networks (GNNs) on large, biologically relevant proteins. DISPEF contains over 200,000 proteins with implicit solvation free energies,
0
3
29
I am excited to share our latest #preprint: ChromoGen: Diffusion model predicts single-cell chromatin conformations https://t.co/h5FORbb0io As the title suggests, we achieved de novo prediction of 3D chromatin structures using DNA sequence and ATAC-seq using AI.
2
8
64
Our explicit ion paper is now online at Elife: https://t.co/4AlISHvEIL. But who has time to read anyway? Luckily, @XingchengLin made an excellent video that you can watch at:
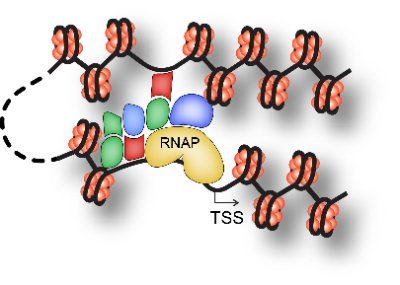
It's as if the very fabric of chromatin's existence is intricately woven through these inherent interactions, shaping its three-dimensional structure. https://t.co/Wl7MQsOv52
0
1
15
Our review on chromatin organization is online at the Annual Review of Biophysics! Spoiler alert, we took a more physical chemist's perspective on the problem. https://t.co/5g7m9CoXOk
0
12
62
Thanks to the efforts of our group members. This new work provides a new role of micropolarity in condensates.
A new paper from Songtao Ye @ye_songtao44002 and Xin Zhang @kfzhangxin @Westlake_Uni reveals the critical role of micropolarity in controlling the structure and miscibility of subcompartments in #biomolecular condensates. Read it here https://t.co/B9SZv0dHhF
3
11
84
Interested in IDPs and biomolecular condensates? Do you ever wonder why IDPs adopt low-complexity domains? How do you map a given protein sequence into stickers and spacers? We attempt to address these questions in our latest preprint.
biorxiv.org
Biological condensates play a vital role in organizing cellular chemistry. They selectively partition biomolecules, preventing unwanted cross-talk and buffering against chemical noise. Intrinsically...
1
4
29
We welcome new postdoc, Joe Paggi, and grad students Camryn Carter and Ivan Riveros to the group. I am very much looking forward to working with you all.
0
0
12
Our paper just appeared online at ACS Cen Sci! Really impressed with the journal's efficiency.
pubs.acs.org
Implicit solvent models are essential for molecular dynamics simulations of biomolecules, striking a balance between computational efficiency and biological realism. Efforts are underway to develop...
I am very excited to share our latest preprint on building transferable CG force fields with GNNs and contrastive learning. We believe the two techniques offer a path to build CG models that rival the accuracy of atomistic models.
1
1
29
I am excited to share our whole nucleus study that provides structural and dynamic modeling of the diploid genome (100kb resolution), lamina, speckles, and nucleoli. We have consolidated our modeling effort into an open-source GPU-enabled software.
biorxiv.org
The intricate structural organization of the human nucleus is fundamental to cellular function and gene regulation. Recent advancements in experimental techniques, including high-throughput sequenc...
1
15
58
Our OpenABC paper just appeared online: https://t.co/i2Or11vfov During revision, we also added multiple new force fields to the software, including the wonderful Mpipi for RNA from @rcollepardo
journals.plos.org
Author summary Biomolecular condensates are essential cellular structures that underpin various cellular processes, including RNA splicing, ribosomal RNA processing, and stress response, etc. These...
Ever wonder about performing simulations of biomolecular condensates but don't know where to start? Wonder no more! Check out our software OpenABC that allows condensate simulations with only a PDB file
1
1
14
I am very excited to share our latest preprint on building transferable CG force fields with GNNs and contrastive learning. We believe the two techniques offer a path to build CG models that rival the accuracy of atomistic models.
biorxiv.org
Coarse-grained (CG) force fields are essential for molecular dynamics simulations of biomolecules, striking a balance between computational efficiency and biological realism. These simulations employ...
0
9
41
Our paper on efficiently inverting Hi-C contacts to interaction energies is out: https://t.co/XPSNmQQE7W. This inversion could facilitate polymer simulations as well as contact feature extraction. Great job, Greg!
0
2
20