BEAP Lab (Biology & Ecology of Abundant Protists)
@beaplab_ibe
Followers
748
Following
152
Media
33
Statuses
100
The Biology and Ecology of Abundant Protists (BEAP) Lab at the Institut de Biologia Evolutiva (CSIC-UPF). Principal Investigator: Daniel J. Richter
Barcelona
Joined April 2014
RT @IBE_Barcelona: 📣Last days to apply!📣. 3️⃣ PhD student positions at the @IBE_Barcelona research groups of @multicellgenome , @beaplab_ib….
0
16
0
RT @Pawel_Burkhardt: "A large colonial choanoflagellate from Mono Lake harbors live bacteria" out now in @mbiojournal. We isolated & charac….
0
22
0
The BEAP lab is recruiting a PhD student (4 years) to start between January-March, 2025!. The project will isolate novel, environmentally abundant protists, and investigate how they live in the lab and in the ecosystem. Apply by: 10 September.More info:
beaplab.org
image credit: C. Sigona
📣 New PhD Student position! 📣. Join @IBE_Barcelona as a PhD Student in the @beaplab_ibe 🧫🔬 to discover the most abundant #protist species in the world, and their role in global #marine #ecology.🦠🧬. 📆 Apply by September 10th. #JoinOurTeam!. ✍️
0
10
12
This was a great collaboration with BEAP Lab member @ungalvez and our colleagues Laurent Guégen, @Pnatsidis and @TelfordLab. (Yes, it was actually published way back in March, but better late than never!).
0
0
0
Our preprint has now been published: Take home message: if you are performing ancestral reconstructions based on sequence homology that only permit a gene/domain to be gained once on a tree (e.g., Dollo parsimony, phylostratigraphy), please use caution!.
academic.oup.com
Abstract. Ancestral reconstruction is a widely used technique that has been applied to understand the evolutionary history of gain and loss of gene familie
New preprint! For years, we had worried that Dollo parsimony, which is based on principles developed for morphological characters, was not an appropriate choice for ancestral gene content reconstruction based on sequence homology. Now we can say: it’s not!
1
3
7
Twitter changed its name, and now we are following suit. Please say goodbye to our previous username, the powerful sorcerer Balkalarre. Please say hello to our new username, the Biology & Ecology of Abundant Protists Lab at the Institut de Biologia Evolutiva (IBE): @beaplab_ibe.
0
0
14
RT @JohanD13016359: Happy to share our new preprint from postdoc @CarolineJuery revealing how the transportome of a symbiotic microalga (P….
0
7
0
RT @multicellgenome: Interested in exploring evolution, cell biology, & ecology of #protists in a Mediterranean setting? 🌊🔬 Barcelona hosts….
0
40
0
RT @IBE_Barcelona: 💥 #11F 💥. At @IBE_Barcelona we celebrate the International Day of Women and Girls in Science with the launch of the #Clo….
0
12
0
RT @fonamental: #Protists double animal's biomass on earth. Despite their importance, our knowledge of these organisms is greatly hampered….
0
48
0
RT @IBE_Barcelona: #Evolution is your passion?🧬🦠🐆. 📢 Check out the open call for @FundlaCaixa #INPhINIT Retaining 2024 and #joinourteam at….
0
6
0
Next, we compared Dollo (A) to ML (B) on a real dataset of Pfam domains. Dollo’s LECA has more domains than any extant eukaryote followed by widespread losses; ML’s has fewer LECA domains and balanced gains/losses. Which to trust? Our results suggest caution with Dollo parsimony.
0
0
2
With @Pnatsidis and @TelfordLab’s simulated sets of 5,000 orthologs with no gains or losses, we tested our suspicion together. We compared Dollo to maximum likelihood (with Laurent Guégen), finding that Dollo consistently overestimates ancestral gene content towards the root.
1
0
0
New preprint! For years, we had worried that Dollo parsimony, which is based on principles developed for morphological characters, was not an appropriate choice for ancestral gene content reconstruction based on sequence homology. Now we can say: it’s not!
2
11
33
The BEAP lab would like to welcome two new members! Xènia Maya joins us as a lab technician for the IBE Microbe Hub (joint with @multicellgenome and @delCampoLab), and Viktoriya Petlovana joins us as a visiting scientist from Taras Shevchenko National University of Kyiv.
0
2
14
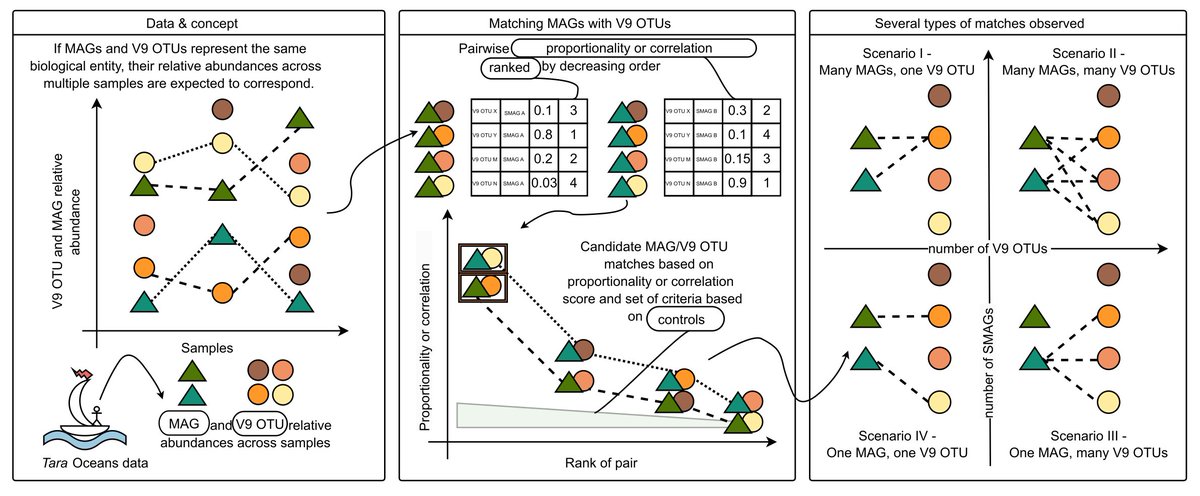
Furthermore, V9 OTUs and MAGs from a given taxonomic group did not necessarily fall into the same scenarios of correspondence, indicating that other factors beyond taxonomic lineage are likely to shape the V9 OTU-MAG relationship.
0
0
0
In some cases, there was a 1-to-1 correspondence between a single V9 OTU and a single MAG, meaning they likely represent the same underlying biological entity. But we also found numerous instances of one-to-many, many-to-one, and many-to-many V9 OTU/MAG correspondence scenarios.
1
0
1
We compared 18S V9 OTUs versus MAGs from Tara Oceans. Due to the way MAGs are constructed, they do not contain 18S sequences. Instead, we developed a method with a series of controls to detect robust correspondences between V9 and MAG relative abundances across samples.
1
0
2
Metabarcodes and metagenome-assembled genomes are valuable approaches to study protist community structure and function. How do they correspond to one another? Our answer: it’s complicated, and it doesn’t necessarily depend on taxonomic group. Preprint:
1
34
85
RT @fburki: A couple lab member Anne and @thealgaeman have been visiting @balkalarre and @RMassanaM labs for learning culturing techniques….
0
2
0