Adam Siepel
@asiepel
Followers
5K
Following
2K
Media
12
Statuses
2K
Computational biologist at Cold Spring Harbor Lab. Evolutionary geneticist. Reformed computer scientist. Sometimes writer. Lapsed programmer. Dad.
Huntington, NY
Joined May 2009
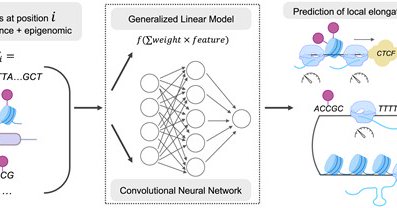
Ling Liu’s beautiful PhD work on modeling transcriptional elongation and its genomic and epigenomic determinants is now published. A technical tour de force with a number of interesting biological observations
academic.oup.com
Abstract. Rates of transcription elongation vary within and across eukaryotic gene bodies. Here, we introduce new methods for predicting elongation rates f
1
3
15
The abstract deadline for ProbGen '25 has been extended to January 24. Turnout has been good so far but we're hoping to increase it a bit more, so there's still time to submit if you have an idea for a talk or poster!.
meetings.cshl.edu
Cold Spring Harbor Laboratory Meetings & Courses -- a private, non-profit institution with research programs in cancer, neuroscience, plant biology, genomics, bioinformatics.
0
4
8
Please note that the abstract deadline for ProbGen '25 is this Fri, Jan 10. Please join us. We have a great line-up of keynote speakers and session chairs.
meetings.cshl.edu
Cold Spring Harbor Laboratory Meetings & Courses -- a private, non-profit institution with research programs in cancer, neuroscience, plant biology, genomics, bioinformatics.
0
7
15
RT @Nowak_Lab: We are looking for a passionate #Postdoc to join our team in NYC @WeillCornell! Interest in #ProstateCancer, #Evolution, #Me….
0
15
0
Latest preprint from our group, on a clever approach devised by Ziyi Mo to address a critical limitation of simulation-based deep-learning approaches in popgen.
biorxiv.org
Investigators have recently introduced powerful methods for population genetic inference that rely on supervised machine learning from simulated data. Despite their performance advantages, these...
2
2
13
Reminder that the abstract deadline for ProbGen '23 is one week from today!
meetings.cshl.edu
Cold Spring Harbor Laboratory Meetings & Courses -- a private, non-profit institution with research programs in cancer, neuroscience, plant biology, genomics, bioinformatics.
0
4
8
RT @david_a_knowles: @asiepel and I are hosting our first post(?)-pandemic in-person pop/statgen seminar tomorrow 4pm at @nygenome with @pa….
docs.google.com
Thursday, November 10, 2022 New York Genome Center, 1st Floor Auditorium 101 Avenue of the Americas, New York, NY 10013 4:00 - 5:00 PM ET - Speaker Presentation and Q&A 5:00 - 5:30 PM ET - Reception
0
2
0
RT @lizzyzhao: We are excited to announce that New York area population genomics meeting is back. The next meeting is on January 27, 2023,….
nyapg23.wordpress.com
Official site for NYAPG 2023
0
18
0
@YixinZhao1989 One of the most fun and satisfying parts of the project was iterating between simulation and model development. Using Yixin's simulator, we kept discovering new aspects of the process to incorporate into the model. 13/14.
1
0
1
@YixinZhao1989 was the lead on this project and did a massive amount of work to bring it over the finish line. Also joint work with Ling Liu, who has a related paper in prep (coming soon!) 12/14.
1
0
0