Anish Simhal
@aksimhal
Followers
302
Following
2K
Media
6
Statuses
345
Postdoctoral Fellow at @MSKCancerCenter Mathematical Oncology Initiative. Researching network science, genomics, oncology. Previously @DukeU, @UVA
New York City
Joined September 2009
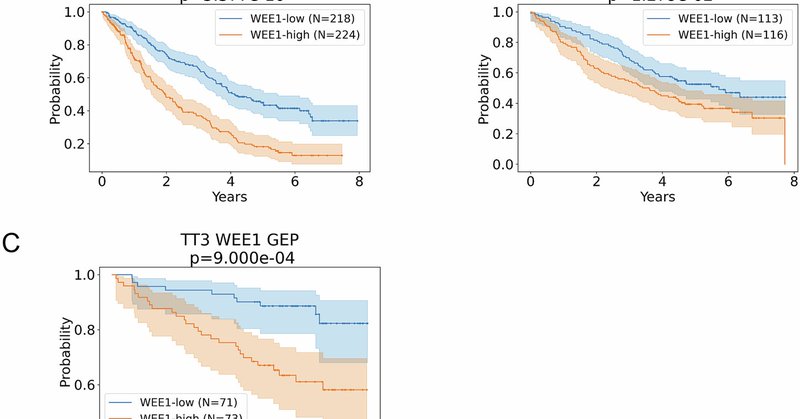
1/ 🚨New research alert! 🚨Our study in @BloodCancerJnl shows that high WEE1 expression is an independent predictor of poor survival in multiple myeloma (MM), with @RossFirestone, @MalinHultcrantz, @LarryNortonMD, @szusmani, and the rest of the @MSKCancerCenter team! 🧵👇.
1
5
16
RT @MSKCancerCenter: #Lungcancer can co-opt genes that normally help a fetus develop and evade the mother’s immune system, according to res….
0
5
0
9/ Want to try ORCO? If you're working with omics data & want to see how network curvature can enhance your analysis, I’d love to help! Please reach out. #Bioinformatics #NetworkBiology #SystemsBiology.
0
0
0
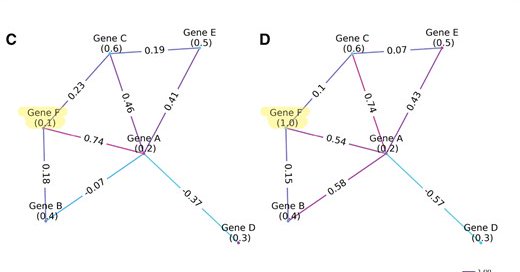
8/ ORCO is open-source & [relatively] easy to use! 🎉 Install via pip and start exploring network robustness in your data. Please reach out if you find any bugs or issues! . 📌 Paper:
academic.oup.com
AbstractMotivation. Although recent advanced sequencing technologies have improved the resolution of genomic and proteomic data to better characterize mole
1
0
0
Link to manuscript: Thanks to @jianlincheng for the comments and editing!.
academic.oup.com
AbstractMotivation. Although recent advanced sequencing technologies have improved the resolution of genomic and proteomic data to better characterize mole
1
0
0
🚀 Just published in Bioinformatics! Introducing ORCO: Ollivier-Ricci Curvature-Omics, a python package for analyzing robustness in biological systems. A 🧵 on what it is, why it matters, and how you can use it. With the great team from @MSKCancerCenter.
1
1
6
RT @oncodaily: Myeloma Paper of the Day, February 22nd, suggested by @Myeloma_Doc .@RossFirestone . #Myeloma #Canc….
oncodaily.com
Myeloma Paper of the Day, February 22nd, suggested by Robert Orlowski / Anish Simhal, cancer, Joseph Deasy., Jung Hun Oh, Kylee Maclachlan, Larry Norton,
0
1
0
RT @Myeloma_Doc: #Myeloma Paper of the Day: WEE1 expression is prognostic independent of known biomarkers, differentiates outcomes associa….
0
8
0
12/ Bottom line:.🔹 High WEE1 = worse survival in MM.🔹 WEE1 is independent of known risk factors.🔹 Targeting WEE1 might be a new therapeutic avenue.🔗 Read more:
nature.com
Blood Cancer Journal - High WEE1 expression is independently linked to poor survival in multiple myeloma
1
1
1
8/ So why is this important?.✅ WEE1 expression could be used as a new prognostic biomarker in MM. ✅ WEE1 inhibitors, already in trials for other cancers (including those by @ZentalisP & @DebiopharmNews) might be a therapeutic option for MM patients.
1
0
0