Murphy Lab
@_Murphy_Lab
Followers
458
Following
20
Media
17
Statuses
50
Structural and mechanistic study of proteins, especially redox- and metalloproteins
Frankfurt, Germany
Joined September 2021
Happy to share that our work on the FDX2-bound core ISC complex is now published in @NatureComms 🎉. By the way, we are now also on @bluesky! 🦋
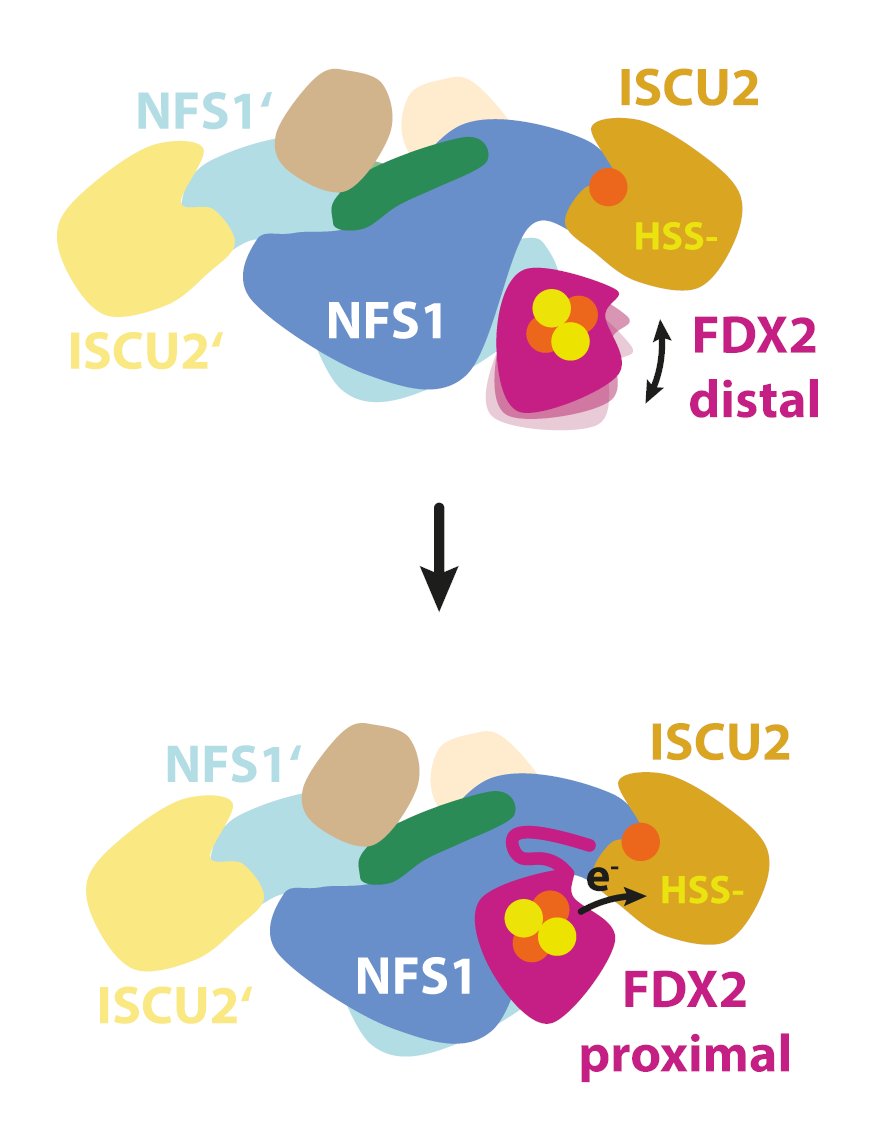
🎉 Our latest preprint is now online 👉 Congrats to @RalfSteinhilper who determined #cryoEM structures of the ferredoxin 2-bound core iron-sulfur cluster assembly complex under reducing and turnover conditions at 2 Å resolution. More below! 👇. 🧵 1/8
0
3
9
Our elemental mapping work, combining cryo-EM and EELS to REEL analysis, is now also published on Nature Methods:. For a highlight “reel”, see here 👇.
Happy to announce that our efforts to establish elemental mapping for cryo-EM ❄️ by EELS 🌈 are now shared on bioRxiv: Congratulations to @zoliviapg and co-authors!. But wait - Can we map elements in 3D?.🧵👇 1/6
1
13
55
Congrats to @MaxYin_211013 and co-authors! Big thanks to X-ray team: Tristan Wagner, Olivier Lemaire and Mélissa Belhamri @MarineMicrobio; computational team: Gerhard Hummer and José Guadalupe Rosas Jiménez @HummerLab; MS team: Anna Shevchenko @mpicbg for the great collaboration!.
0
0
4
Happy to see our study on the structure of 46-kDa Streptococcus pneumoniae NOX out in @NatureSMB! Congratulations to the authors @PABLOSANSEGUND3 and @vicsnorr 🥳.Check it out here (open-access) 👉
Excited to share our preprint: “Structural and mechanistic insights into Streptococcus pneumoniae NADPH oxidase” in which we obtained a structure of a 46-kDa membrane protein to 2.2 Å resolution without fiducials.
0
9
37
#Mitochondria are known as the ‘powerhouses’ of the cell. But an even more widely conserved function is the biosynthesis of inorganic protein cofactors called iron-sulfur #FeS clusters, which are built de novo from free cysteine and iron by the core ISC complex. 🧵 2/8.
1
0
1
🎉 Our latest preprint is now online 👉 Congrats to @RalfSteinhilper who determined #cryoEM structures of the ferredoxin 2-bound core iron-sulfur cluster assembly complex under reducing and turnover conditions at 2 Å resolution. More below! 👇. 🧵 1/8
2
15
94
A big thanks for their contributions, especially to @_Higor_Rosa, as well as Dietmar Riedel, @YuSebyChen, @FilipPetegem, and the always-helpful team from CEOS whose awesome CEFID we have been relying on, as well as @DECTRIS_News whose ELA has been crucial to this project. 🧵 5/6.
2
0
9