Yanqiao ZHU
@Zhu_Yanqiao
Followers
930
Following
818
Media
9
Statuses
221
CS PhD @UCLA | AI for Science, Autonomous Scientific Discovery
Los Angeles
Joined June 2020
What kind of data should we prioritize during self-training? Confident ❌ Uncertain ✅ We’re excited to introduce 🤔EAST 😎— a novel weighting strategy that prioritizes uncertain data during self-training. EAST uses a mapping function with a tunable sharpness parameter to
4
42
197
Mitigating racial bias from LLMs is a lot easier than removing it from humans! Can’t believe this happened at the best AI conference @NeurIPSConf We have ethical reviews for authors, but missed it for invited speakers? 😡
181
803
4K
1/3 Today, an anecdote shared by an invited speaker at #NeurIPS2024 left many Chinese scholars, myself included, feeling uncomfortable. As a community, I believe we should take a moment to reflect on why such remarks in public discourse can be offensive and harmful.
180
565
4K
🚀🚀🚀Want to develop a cutting-edge video generation model towards Sora? Please dive into Apple’s latest recipe and studies for scalable video generation models🔥🔥🔥. In this work, we aim at providing a transparent and detailed recipe 📖 for model architecture, training
9
46
131
A big THANK YOU to each and every one of the participants, presenters, tutorials, and local organizers for making the third @LogConference possible! 💙
0
6
27
I will give a talk at the @LogConference on "Integrating Graph Neural Networks and Large Language Models". The conference is virtual, free to attend, live-streamed, and recorded. https://t.co/PwtgTz0Z4a Hope to see you there!
2
44
164
😄I did a brief intro of RLHF algorithms for the reading group presentation of our lab. It was a good learning experience for me and I want to share the github repo here holds the slides as well as the list of interesting papers: https://t.co/TFIcpwUqul Would love to hear about
7
27
259
📢New LLM Agents Benchmark! Introducing 🌟MIRAI🌟: A groundbreaking benchmark crafted for evaluating LLM agents in temporal forecasting of international events with tool use and complex reasoning! 📜 Arxiv: https://t.co/ikuRg2SQtr 🔗 Project page: https://t.co/hrMjFRk6gR 🧵1/N
14
70
301
Check out our new work in LLMs for molecule optimization!
🧵1/n LLMs significantly improve Evolutionary Algorithms for molecular discovery! For 18 different molecular optimization tasks, we demonstrate how to achieve SOTA performance by incorporating different LLMs! Learn more in our new paper! Website: https://t.co/S0zw97Ialr(w/ Code)
0
0
3
How to control LLM behavior with LLM-as-a-judge? Check our paper: "Self-Control of LLM Behaviors by Compressing Suffix Gradient into Prefix Controller" Website: https://t.co/PZoWUqrAHN Paper: https://t.co/Xwl8wEJX0g Code: https://t.co/x0yiK77qaW
2
13
47
📢 New paper alert! Introducing STIC (Self-Training on Image Comprehension) that enhances the understanding and reasoning capabilities of LVLMs through self-generated data 🌟 📄 Read the paper: https://t.co/Pzwj5gCIZq 🔗 Project page: https://t.co/y4GRQyKwii 💻 GitHub Repo:
1
44
200
Our SciBench paper got accepted to #ICML2024! 🔬We benchmarked leading open-source & proprietary LLMs, including multimodal models, on the updated SciBench Check out the paper at
arxiv.org
Most of the existing Large Language Model (LLM) benchmarks on scientific problem reasoning focus on problems grounded in high-school subjects and are confined to elementary algebraic operations....
🧸We introduce SCIBENCH, a challenging college-level scientific dataset designed to evaluate the reasoning abilities of current LLMs (#gpt4, #chatgpt). 🐻We find that no current prompting methods or external tools improves all capabilities. Github: https://t.co/JXfGuDiJi7
1
6
33
6⃣ Check out MARCEL in Session 4 on May 8. MARCEL is a benchmark that comprehensively evaluates molecular conformations via 4 datasets covering diverse molecule- and reaction-level properties for drug discovery and enzyme design: https://t.co/wEvuFs9Oq5
#ICLR2024
1
3
8
🧬 Still using BM25 for biomedical retrieval? Try out BMRetriever! 🔍 Our new series of retrievers enhance biomedical search with various scales (410M-7B). 🔓 Model/Data: https://t.co/hJkgKZJKcB 🌠 Github: https://t.co/Jkap1bJrOP
#BiomedicalResearch #LLM #Retrieval #OpenScience
github.com
[EMNLP 2024] This is the code for our paper "BMRetriever: Tuning Large Language Models as Better Biomedical Text Retrievers". - ritaranx/BMRetriever
1
11
37
We started releasing the first chapters of our Geometric Deep Learning book and the accompanying slides from the corresponding Oxford and Cambridge courses.
After 3 years, it's time for us to start sharing the chapters of the GDL book! ❤️ Also included: companion slides from our @Cambridge_Uni & @UniofOxford courses 🧑🎓 Chapter 1 is out **now**! More to follow soon 🎉 https://t.co/g7SqyZBCgX 📖 @mmbronstein @joanbruna @TacoCohen
10
104
665
🚀 Great news! The @LogConference 2023 proceedings are now available on PMLR: https://t.co/exSXOOGLo8 — thanks to the entire community! We also have some exciting updates about the next edition of LoG, coming soon... ⌛️
0
15
94
Great work on scaling pretrained GNNs for molecular graphs! Our previous work ( https://t.co/RFB0UBiCh8) also studied the neural scaling behaviors by analyzing the impact of data quantity and quality on molecular representations.
arxiv.org
Molecular Representation Learning (MRL) has emerged as a powerful tool for drug and materials discovery in a variety of tasks such as virtual screening and inverse design. While there has been a...
On the Scalability of GNNs for Molecular Graphs "Scaling laws for GNNs" This is about activity/property prediction: - Pre-training on more molecules helps When you read the paper, keep in mind: SOTA for most tasks is descriptors+xgboost (Fig 4). P: https://t.co/ZajR1LfWuP
0
0
13
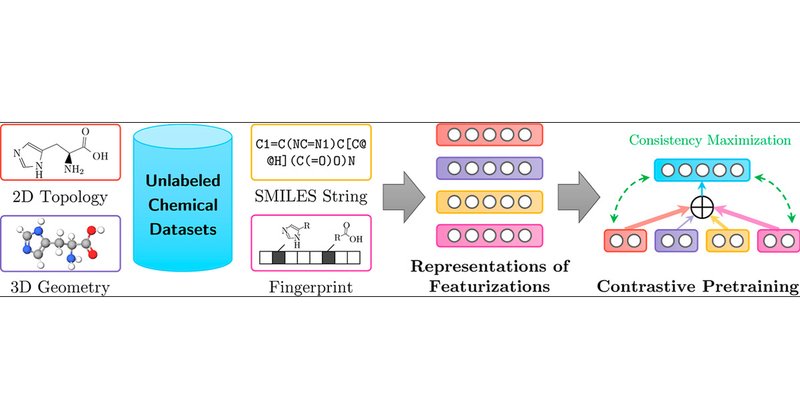
Molecular Contrastive Pretraining with Collaborative Featurizations https://t.co/pe0rynh3af
@Zhu_Yanqiao @YuanqiD
#JCIM Vol64 Issue4 #MachineLearning #DeepLearning
pubs.acs.org
Molecular pretraining, which learns molecular representations over massive unlabeled data, has become a prominent paradigm to solve a variety of tasks in computational chemistry and drug discovery....
0
5
25