Brett Phinney
@UCDProteomics
Followers
5K
Following
6K
Media
553
Statuses
6K
Director: UC Davis Genome Center Proteomics Core. Analyzer of weird samples . Proteomics Radio Hour crooner. Currently twitterpated
UC Davis Genome Center
Joined March 2009
If anyone's interested we made about 20 proteomics videos/lectures/tutorials mainly for grad students post-docs etc interested in proteomics
23
183
513
Don't forget to register for this upcoming hybrid seminar, join us Nov 6 at 10am-12pm either on Zoom or in-person at GBSF, Auditorium.
Nov 6: Join our guest lecture seminar featuring two great speakers! Prof. Worsnop on organics in aerosols at 10am, and Prof. Kuo is focused on pharmaceutical analysis. Hybrid event. Join here: https://t.co/siuq5vhNRa
#Lipidomics #Metabolomics #Aerosols
0
1
11
This was a little reels video we made for the @genomecenter Halloween symposium this year. Btw we do know what RNA is , but it was too fragile and got cut 😉 https://t.co/ezYaV9kAAv
instagram.com
1
0
10
Seems like a good time for a list some of my favorite truly #OpenSource #proteomics tools. In no particular order: 1. Comet - The Sequest algorithm for everyone.
github.com
An tandem mass spectrometry (MS/MS) sequence database search tool. - UWPR/Comet
1
13
73
DIA-NN 1.9.2 is out! There are some major improvements, please check the release notes
github.com
DIA-NN 1.9.2 is a major update with several key performance and functionality improvements. Notes DIA-NN 1.9.2 is a free Academia-only version. Our spin-off Aptila Biotech is now preparing an ente...
5
22
99
These 2 I consider required reading. I have read them over 2-3 dozen times over the last decade or 2 . yes I'm old now. I didn't used to be. how did that happen? https://t.co/Rz9adGitVX
https://t.co/QMvi6c38US
4
0
16
Happy to hear I'm wrong, but man I would not spend my energy on SILAC labeling these days compared to the alternatives. Even MS1 LFQ DDA with MBR on non silac samples seems better to me. that works a lot better now than it used to.
1
0
3
SILAC DIA shows promise but still I think you are far better off using that time and energy for more replicates than SILAC labeling
1
0
3
I don't think the advantages of combining your samples far upstream is a real advantage any more like it used to be. I think you are far better off using sample prep automation to decrease variation and having more replicates
1
0
3
I think MS1 quant is just not as good as DIA MS2 level quant for most of the S/N range. You have far less curves to work with & at low S/N peak picking is seems far worse. Personally, I think combining them is the way to go which you can't do with SILAC DDA.
1
0
2
1. Designing a proper experiment that is not underpowered seems really tricky.
1
0
1
As I go through a recent SILAC experiment I'm becoming convinced it's a relic that should probably be discarded.
1
0
12
Way to go Thermo , another fine example of packaging. You know people with degrees in packaging do exist, maybe you should think about hiring some of them ? . https://t.co/PzFGeIa8uV
2
0
21
Received this with my evotips today. I wish I had a cute very tiny wizard in the lab! I feel like I’d pat his head for good luck , but he would always be grumpy because something was always broken
0
0
14
I almost never analyze SILAC samples anymore. Is it becoming less popular? Anyway I was really impressed with using the latest version of Fragpipe -w/ skyline integration for dda and Spectronaut 19 for DIA.
4
0
27
0
0
0
Workflow = S-trap -> timsTOF HT -> diaPASEF -> Spectronaut 19 . Easypeasy
0
0
3
Worlds first Pancake Proteomics? Could be!! We analyzed the pancakes we will be serving at our next @genomecenter coffee. Here are the top proteins you will be eating. We identified about 6K total. Credit to @DixonLydixon & Michelle in the lab
4
2
19
What is the appropriate way to compute FDR when you have a low number of peptide identifications. <50 or <100 or <500. The standard target/decoy formula works well for a few 1000 ids but is harsh for a few dozen. Side note, I don't really know if FDR matters for low numbers...LOL
2
1
5
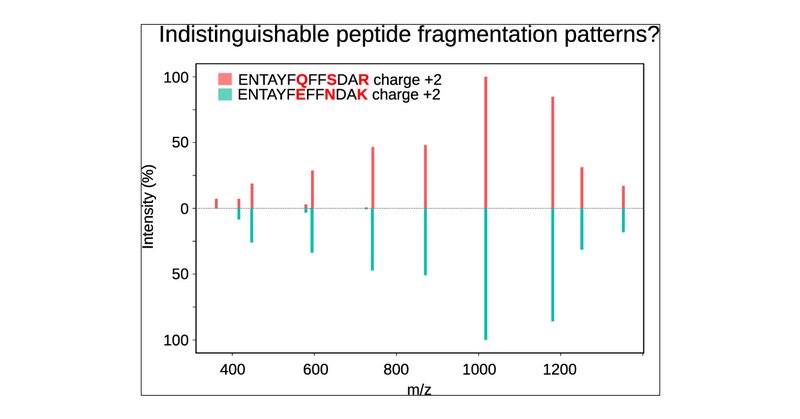
Ever wondered how many closely eluting peptides have the same fragmentation pattern? In collaboration with @wilhelm_compms we explored the theoretical nonaccessible portion of the human proteome. Check our new publication at @an_chem #proteomics
pubs.acs.org
Mass-spectrometry-based proteomics has advanced with the integration of experimental and predicted spectral libraries, which have significantly improved peptide identification in complex search...
0
10
37
There are still a few more days till Bits and Bites #7: Using MetaboAnalyst for Metabolomics Statistics and Data Visualizations. If you are interested sign up today! 📅 Date: October 3, 2024, 9am–1pm PT 👨🏫 Instructor: Dr. Jianguo (Jeff) Xia 🔗
metabolomics.ucdavis.edu
Bits & Bites # 07: Using MetaboAnalyst for Metabolomics Statistics and Data VisualizationsDate: October 3, 2024Instructor: Prof. Jeff Xia, McGill UniversityRegistration & Payment: here
1
4
6