The LOTUS Initiative
@TheLOTUSInitia1
Followers
184
Following
43
Media
3
Statuses
120
[email protected] https://t.co/YYE4LOPjYS
Joined May 2021
The #LOTUS initiative article is finally officially published at @eLife ( https://t.co/bSh8P48syS). This is an international effort to make #naturalProducts knowledge #open, #collaborative, and #machineInterpretable. (a 🧵) (1/7)
elifesciences.org
The LOTUS initiative builds, through the Wikidata knowledge graph, a virtuous cycle of data sharing practices for natural products research.
3
33
57
#IDSM (the small-molecule database developed by @ELIXIRCZ) now also allows you to search for small molecules by their mass spectrum via #SPARQL. https://t.co/RQmU4WeymX
academic.oup.com
AbstractSummary. The Integrated Database of Small Molecules (IDSM) integrates data from small-molecule datasets, making them accessible through the SPARQL
0
5
9
Our Prague #compMS Workshop hosted 100 researchers +online. It was a great success thanks to all presenters, participants, sponsors, and venue @IOCBPrague. @mzmine_project #SIRIUS_MS #matchms @wikidata @TheLOTUSInitia1 @GNPS_UCSD #massspec #metabolomics 1/n ...
2
16
79
The two last papers from the Wolfender lab ( https://t.co/0xi3XaoleB and https://t.co/fXTVpVVZfu
@LuisQuirosG) shared the data about their beautiful new natural products (some named in honor of @GisEugenia) with the community! 🥳Don't be shy, just do it! #shareYourSMILES
0
3
8
Actually, it is a privilege to be so closely associated to the Lotus initiative... because it is yet another autonomous achievement directly supported by the TNA arm of #BICIKL ! @plazi_ch @ISBSIB @hes_so ETH_en #bicikluberalles
Great to be hearing more about the collaborations between members of the @Bicikl_H2020 consortium and external stakeholders like the Swiss startup @k8pixels at the #BiCIKL_H2020's #FinalGA. 📸Earlier today, we heard from @Adafede (LOTUS) about further uses of #nanopublications!
0
1
4
Interesting discussion on how Lotus and chEBI could contribute to build a strong ecosystem for small & natural molecules ! @ISBSIB @ETH_en @emblebi @ELIXIREurope @hes_so @wikidata @Adafede
@Pensoft @Bicikl_H2020 @ELIXIREurope @patrickruch @AcaciaJoe @myrmoteras @cabbageleek @rmwaterhouse @DimitrisKoureas @plbuttigieg @pmergen @deb_caucheteur Delighted to welcome and catch up with @Bicikl_H2020 colleagues at the #finalAGM in Cambridge/Hinxton. Much achieved and still things to look forward to.
0
1
4
The #BiCIKL_H2020 morning at #finalga in #cambridge continues with Anke Penzlin (@geobiodiversity) https://t.co/2k6YDJuera and Adriano Ruiz who put together @plazi_ch @ISBSIB and Knowledge Pixels for his project: https://t.co/Uf0wP8jEp2
0
3
4
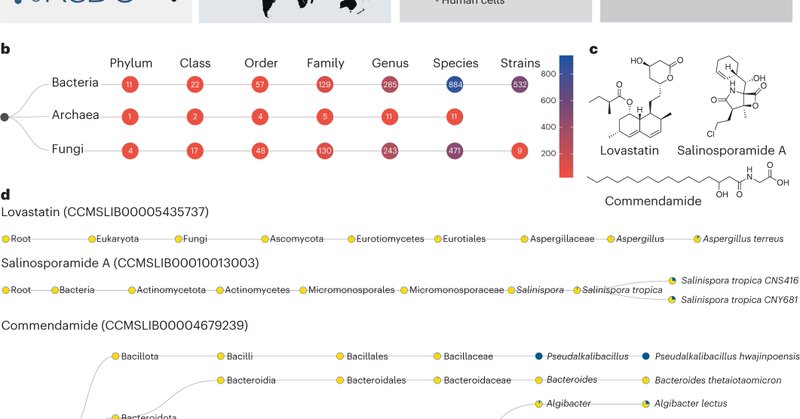
#microbeMASST is finally out! Use the web interface or our Python script to search your (un)known mass spectra against all public MS2 and map the results to taxonomic trees of potential source organisms. 🧵1/n https://t.co/9EwMeBpGIe
nature.com
Nature Microbiology - microbeMASST is a tool to associate known and unknown metabolites to microbial producers leveraging untargeted metabolomics data.
2
33
94
congratulations to @Adafede and @TheLOTUSInitia1
President of the ORD Prize Jury @EvieVergauwe presents the Gold Winner of the #ordprize23: it‘s „The LOTUS Initiative“ by @Adafede! Congratulations 🎉
0
2
3
@TheLOTUSInitia1 It also relies on multiple other wonderful softwares, each adding its own touch of ingenuity to the final result. Hats off to everyone involved in the development & maintenance of this software! 🎩 🎉 @mzmine_project @GNPS_UCSD @boeckerlab @bright_giant @jo_rainer et al. 3/5
1
1
0
This approach builds on the tools and resources we developed earlier to integrate prior knowledge into our workflow ( https://t.co/mRkVXVkaWC and https://t.co/5Svz8fpY6D). 🪷 @TheLOTUSInitia1 2/5
elifesciences.org
The LOTUS initiative builds, through the Wikidata knowledge graph, a virtuous cycle of data sharing practices for natural products research.
1
1
2
Very happy to share the 5th chapter of my thesis now in open access: https://t.co/gNPXCNLWsB
@JAgFoodChem @ACSPublications We've added new information to our untargeted metabolomics MS analyses 📈: Online semi-quantitative analysis based on Charged Aerosol Detection: 🧵 1/5
2
7
29
Want to make knowledge about #NaturalProducts accessible to everyone in more languages? Help us translate our project page: https://t.co/f9WwzEV85H
#LinkedOpenData #FreeKnowledge @wikidata
0
6
5
We at @Bright_Giant offer #software services #SaaS based on #SIRIUS_MS, #CSIFingerID and #CANOPUS_MS, the leading tools for #untargeted #metabolomics and #NaturalProductDiscovery.
0
2
5
Published on @eLife, @TheLOTUSInitia1 first article is an international effort to make #naturalProducts knowledge #open, #collaborative, and #machineInterpretable. @ISPSO_UniGe Original article ➡️ https://t.co/YiC8hYpiXP
0
9
11
@egonwillighagen @wikidata Awesome! Let me know if we can supply more information here... We have curated some more metabolites in the meanwhile #celegans #lipidomics #metabolomics
0
2
3
As an example of how added masses can be used in a #MS-based search for #naturalproducts, the following query https://t.co/FX8aUy3Kvn returns all compounds corresponding to an exact mass of 524.1765Da ± 10 ppm, along with the organisms in which they are found!
0
0
3
As part of our initiative ( https://t.co/bSh8P48syS), we are pleased to announce that we have recently added over 160,000 CIDs and over 260,000 masses to @wikidata. This will improve interoperability with @pubchem, tell us what you want next! 🥳
elifesciences.org
The LOTUS initiative builds, through the Wikidata knowledge graph, a virtuous cycle of data sharing practices for natural products research.
2
12
19
GNverifier and GNfinder are now at the stable v1.0.0, use brew, github downloads to update them. API changed from /api/v0 to /api/v1
0
2
1