Talkowski Lab
@TalkowskiLab
Followers
1K
Following
95
Media
5
Statuses
120
Understanding the consequences of genomic variation in human disease.
Boston, MA
Joined January 2017
Congrats @Philip_M_Boone on receiving the ASHG Award for Outstanding Early Career Publication! He's a clinical geneticist and Instructor in my lab studying the impact of mutations in cohesin complex genes on genome organization and a cluster of syndromes known as cohesinopathies.
1
5
39
#AGBTPH Kyle Fahr and team @illumina continuing to advance AI methods for variant interpretation - compelling data moving from missense and splice site variants to the challenge of the noncoding genome. Great talk!!.
Day 2 at @AGBT Precision Health 2024 brings another group of incredible speakers including Kyle Farh from @illumina, @aaronquinlan from @UofUGenetics and @ontowonka from @CUSystem. See you there! #AGBTPH
0
0
6
Thanks to all the opening session speakers, the #AGBTPH organizing committee, my co-chairs @NHGRI_Director and @WendyKChung, and as always the #AGBT team for welcoming us to Denver. Looking forward to the rest of the week!!.
0
0
4
Fantastic start to #AGBTPH. Perfect lineup of Nancy Cox on heritability of lab values and biomarker studies, @jorsmo on remarkable progress in neuropsychiatric genetics and suicide prevention, @EimearEKenny fabulous studies of diversity in BioMe, and abstract speaker @LimdiNita.
START ME UP! Co-Chairs @NHGRI_Director from @genome_gov and Mike Talkowski from.@MassGeneralNews and @TalkowskiLab take the stage and kick off Day 1 at AGBT Precision Health 2024. #AGBTPH
1
0
13
Looking for ~1.2M structural variants across diverse populations 👀. Our latest @gnomad_project SV datasets are now released from exome and genome sequencing 🥳. If you are here at #ASHG23 the great @JFuBiostats will give a talk today (11/2) at 1:45pm in Rm. 202A.
As part of #gnomAD v4, in collaboration with the @TalkowskiLab, we have released 1,199,117 genome SVs and 66,903 rare exome CNVs. These data represent the first gnomAD SV dataset released native to the GRCh38 reference genome. (1/2)
0
0
23
Huge congrats to @NancyGenetics who received the @GeneticsSociety Leadership Award. So well deserved!!
0
0
11
The new @gnomad_project release is finally here - well ahead of ASHG👏🎉 . It has been a monumental effort from to bring this resource to the field. More to come on structural variants later but for now - huge congrats and thank you to all members of the amazing gnomAD team!!.
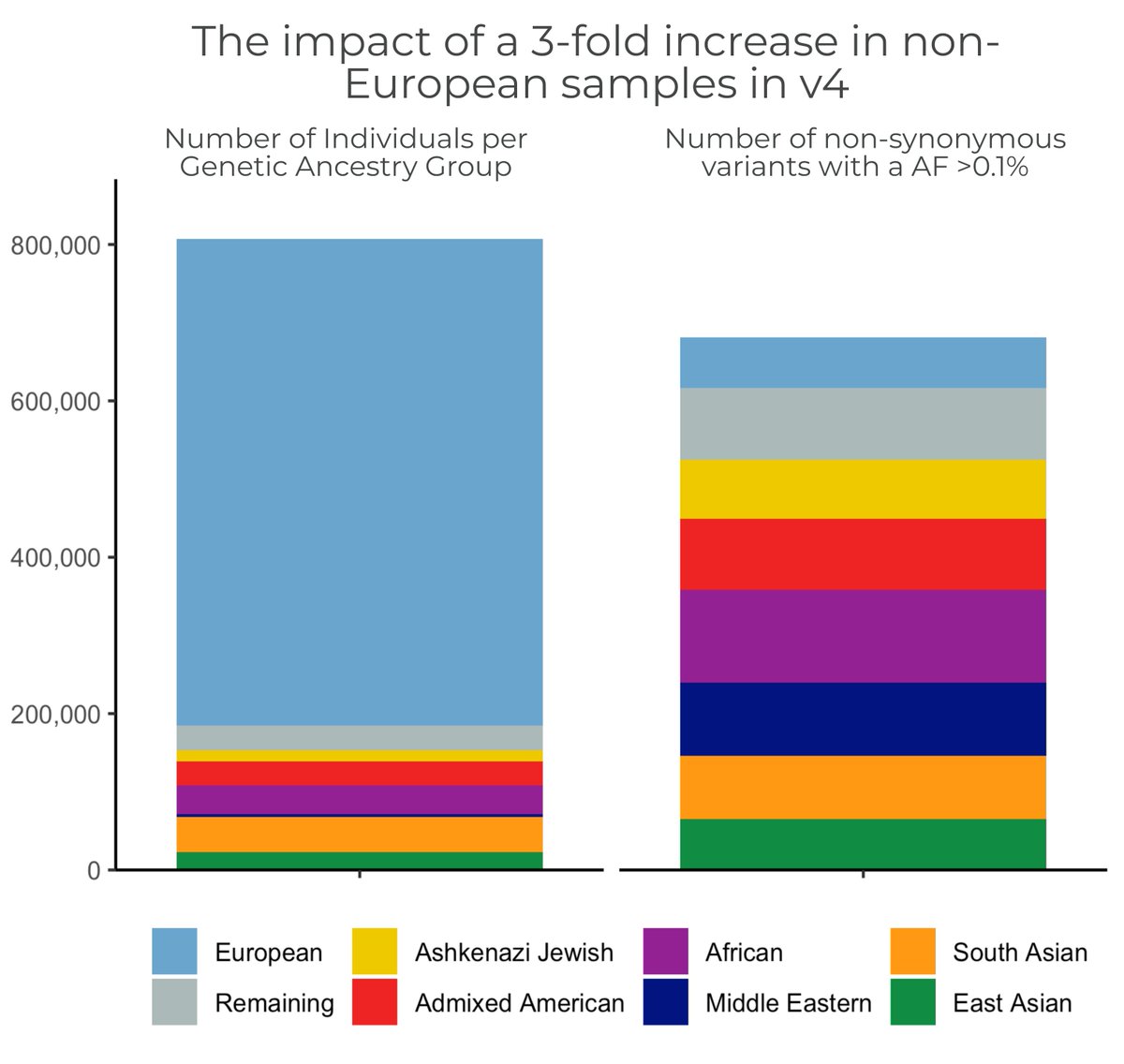
The #gnomAD team is proud to announce the release of gnomAD v4! The v4 dataset includes 730,947 exomes & 76,215 genomes, which is ~5x larger than the v2 & v3 releases combined, & includes nearly 120K indivs of non-European genetic ancestry #ASHG23 (1/11)
0
13
42
RT @CGM_MGH: Happy to announce this year's call for applications supporting our postdoc fellows for the NHGRI-funded MGB T32 Training Progr….
0
7
0
RT @CGM_MGH: Many thanks to the entire CGM community and amazing organizers @GrishchukL, Alysa Doyle, @professorsoukas and Piper McCabe for….
0
1
0
Huge thanks to all GCs and special thanks to our amazing gnomAD scientific operations manager and council member @sambaxtercgc. #gcad2022.
Today is Genetic Counselor Awareness Day and gnomAD would like to recognize all the genetic counselors around the world working in clinics, labs, research, education and beyond. A special thank you to all our GC users and team members! #genechat #GCAD2022
1
1
7
RT @CGM_MGH: The CGM proudly supports all of our Genetic Counselors today on #GeneticCounsellorAwarenessDay!!. The day is dedicated to rais….
0
2
0
RT @ksamocha: MT for Rachita Yadav (@RYbioinfo) #ASHG22: Causal mechanism was alternative splicing of exon 32 + intron retention. TAF1 expr….
0
1
0
RT @HeidiRehm: OK folks, it’s time to start looking at the non-coding portion of the genome! We hope you’ll make use of our newest addition….
0
28
0
RT @CGM_MGH: Check out new paper in @AgingCell from @MGH_CGM associate director, @professorsoukas and his lab. @MGH_RI @harvardmed @Harvard….
0
2
0
RT @NobelPrize: BREAKING NEWS: .The 2022 #NobelPrize in Physiology or Medicine has been awarded to Svante Pääbo “for his discoveries concer….
0
13K
0
RT @PGCgenetics: Congrats to Dr Alicia Martin @genetisaur, who is instrumental in developing statistical methods and resources for multi-et….
0
5
0