Sebastian May-Wilson
@SebastianMayWi1
Followers
169
Following
376
Media
24
Statuses
520
Population genetics PostDoc at the FIMM | PhD from Uni of Edinburgh | Admin at https://t.co/F6S52gwUaC | Will have to remember to not only use this for memes
University of Helsinki
Joined November 2019
RT @ProteinAtlas: A study in Genome Biology presents an integrated resource mapping gene expression across the body, using both single cell….
0
2
0
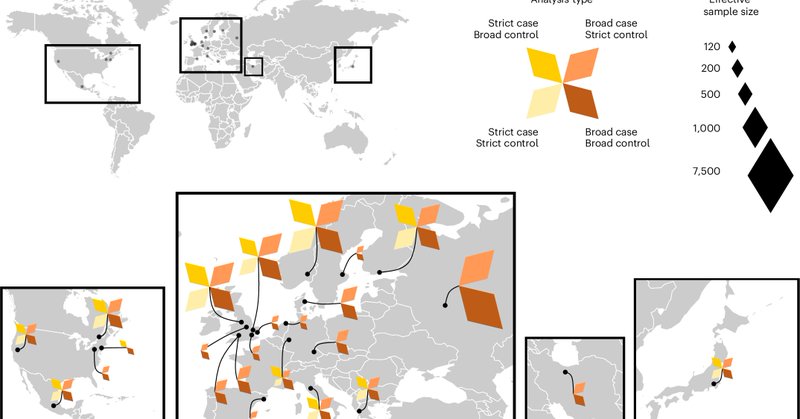
RT @MariosGeorgakis: It's becoming clear that assessing the effects of genetic variants on gene expression within individual cells (sc-eQTL….
0
28
0
RT @DFusco17: #ESHG2025 it’s a wrap! So glad to have joined and caught stunning human genetics highlights 🧬.@eshgsociety .
0
1
0
First time at #eshg2025 and it was an extremely excellent time. Hopefully will see many of your faces again in future. Naturally, I was most impressed by my own work, but what else is new?
0
0
9
Extraordinarily deserved!.
Thrilled to receive the Early Career Award for my presentation at #ESHG2025! 🧬.Big thanks to all the amazing collaborators on this project — couldn't have done it without you. Grateful to the organizers and inspired by the science shared this week!.
0
0
1
The Ganna lab is out in full force at #eshg2025 starting with Zhiyu's talk this morning. If you're interested in the genetic basis of healthcare costs come by my poster tomorrow at 4pm!. Or come by when I'm not there to avoid the awkward part where I stare at you the whole time.
0
0
6
RT @BrentRichards19: I didn't have much hope that we'd find genetic determinants of long-COVID, but after years of work, @VilmaAho, @tomoco….
nature.com
Nature Genetics - A genome-wide study by the Long COVID Host Genetics Initiative identifies an association between the FOXP4 locus and long COVID, implicating altered lung function in its...
0
16
0
RT @Rbn_Hfmstr: 🎉 Hi #ESHG2025, the Statistical Genetics Group and alumni (@zkutalik ) is coming in force!. Check out our latest work on hu….
0
4
0
RT @tuomohartonen: Did you know that some genetic males have plasma proteomes similar to those of genetic females and vice versa? What are….
0
3
0
RT @zkutalik: Very excited to give a virtual talk as part of the @eshgsociety webinar series tomorrow at 4pm (CEST). If you want to listen….
0
7
0
RT @DFusco17: Finally out!🤩.My first first-author publication is now online. We explored the molecular mechanism underlying the genetic cor….
journals.plos.org
Author summary Obesity is linked to many chronic diseases and its prevalence worldwide is increasing. Susceptibility to obesity is known to be due, to some degree, to genetic factors, and such...
0
5
0
RT @doctorveera: Beautiful GWAS study of body mass index (BMI) from deCODE identifying a new risk gene FRS3 associated with BMI with suppor….
0
18
0
RT @andganna: UK launches a national health data service agency.
wellcome.org
Wellcome is partnering with the UK Government to establish a £600 million health data research service. It will simplify secure access to health data for research.
0
2
0
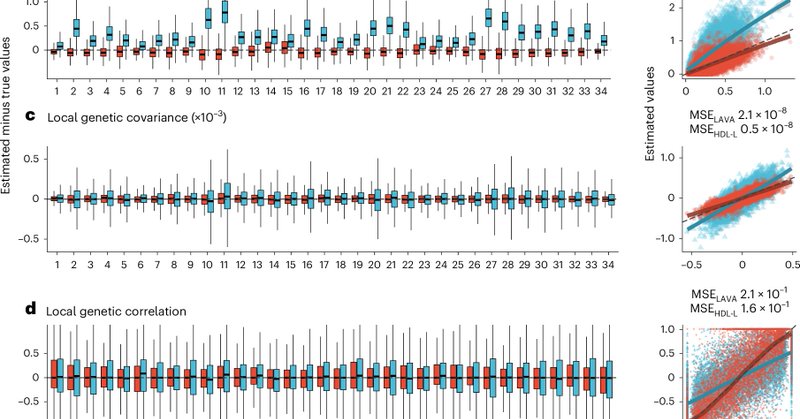
RT @xiashen: Our new method HDL-L, high-definition likelihood for local genetic correlation analysis, is now published in @NatureGenet: htt….
nature.com
Nature Genetics - HDL-L is an extension of the high-definition likelihood method that enables local heritability and genetic correlation analysis with higher accuracy and computational efficiency...
0
12
0
RT @MariosGeorgakis: Polygenic risk scores (PRS) are mainly benchmarked for incident disease in population studies🧬📈. If they start being r….
0
9
0
RT @MariosGeorgakis: Gene expression is regulated at cell level. However, most eQTL studies use bulk-tissue RNAseq. scRNAseq is still ver….
0
23
0
UKB about to do proteomics of 5k proteins in the full 600,000 individuals from the cohort. Genuine ten-fold increase in scale from the previous largest proteomics study (which was also UKB). It's gonna be SO MUCH DATA GUYS.
Today we’re launching the world’s most comprehensive study of proteins circulating in our bodies, which will transform research into new medical tests, drugs and treatments. Discover more:
0
0
0