Lennart Randau
@RandauLab
Followers
196
Following
19
Media
0
Statuses
14
Our laboratory studies Class I CRISPR-Cas biology and archaeal RNA processing pathways at the Philipps-Universität Marburg @Uni_MR. All views are our own.
Joined April 2022
Thank you for the detailled overview of this team effort and your hard work @patrick_pausch! If you want to see beautiful Type IV effector complexes in action, check out:
nature.com
Nature Communications - Type IV-A CRISPR-Cas systems diverge from the general CRISPR-Cas mechanism. To understand this system, the authors determine cryo-EM structures of two evolutionarily...
0
4
13
RT @SelinaRust: Happy to share our article, published today in NAR:. Congrats to @mariana_sanlo24, @RandauLab, @Th….
academic.oup.com
Abstract. Type IV CRISPR–Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) effector complexes are often encode
0
7
0
RT @biowisskomm: "We're CRISPRn our way through the apocalypse". Details can be found at All events are free of ch….
crispr-whisper.de
Übersicht Datum Uhrzeit Veranstaltung Ort 1. Nov. 13:00 – 17:00 Laborkurs 1 „Selber CRISPRn“ K05, Karl-von-Frisch-Str. 8Raum -1202 (s. Lageplan) 1. Nov. 19:00 Abendvorträge K10, Karl-von-Frisch-Str....
0
3
0
Happy to share our paper on archaeal mRNA caps and their processing in @NatureComms: Thank you very much to our very helpful partners in the labs of @JaeschkeLab, @HoerferLab, Jörg Soppa and Nicole Paczia. @Bio_UM, @SYNMIKRO.
0
14
39
Our lab has an open #Phdposition for students interested in RNA and Archaea:.Please feel free to share if you know (or are :-)) a good candidate.
2
21
26
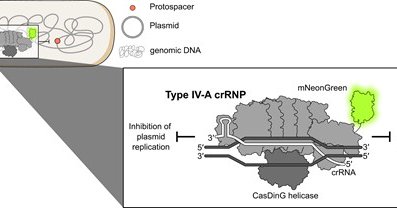
RT @FabBenz: Do plasmids fight other plasmids? 🧵. So happy our preprint is out!💥And yes indeed, we uncover that type IV-A3 CRISPR-Cas targe….
biorxiv.org
Type IV-A CRISPR-Cas systems are primarily encoded on plasmids and form multi-subunit ribonucleoprotein complexes with unknown biological functions. In contrast to other CRISPR-Cas types, they lack...
0
50
0
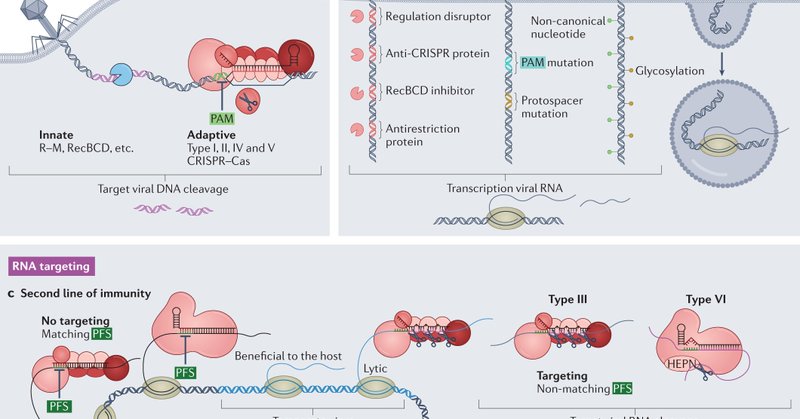
RT @StanBrouns: When microbes have to resort to the more rigorous and diverse RNA targeting CRISPR-Cas strategies to buy time, go into dorm….
nature.com
Nature Reviews Microbiology - In this Review, Brouns and colleagues discuss our current understanding of RNA-targeting type III and type VI CRISPR–Cas systems by detailing their composition,...
0
18
0
RT @GBM_eV: 📢Save the date: GBM and @BioPharmaCL_GER teamed up organizing a half-day event on "The World of RNA - Translation of Basic Rese….
0
2
0
Thank you very much to @petergm24 lab and everybody involved! If you would like to read about how Type IV #CRISPR regulates host gene expression @NatureMicrobiol, please feel free to follow this SharedIt link:
2
14
40
RT @AprilPawluk: can’t wait to review abstracts for @crispr2022 soon! we have lots of short talks available, so be sure to submit your abst….
0
7
0
Great story and we were happy to help a bit with the snoRNA-seq part:.Emergence of the primordial pre-60S from the 90S pre-ribosome: Cell Reports
cell.com
Ismail et al. report the isolation of a primordial pre-60S particle, composed of a specific set of assembly factors and snoRNAs. This assembly intermediate is visualized by cryo-EM while emerging...
0
1
5
Thank you to everybody involved and for all the help from @GBM_eV. It was a really fun and productive meeting! I noticed that we are the only lab lacking any Twitter connections and took this as a hint to finally join in.
A big thank you to the scientific organizers of our #MosbacherKolloquium (f.l.t.r.): Cynthia Sharma @SharmaLab1, Lennart Randau, Gunter Meister @MeisterLab, Markus Landthaler @landthaler_m and Mark Helm @AttheHelm3! What a great face-to-face meeting after a two year break 🤩
0
0
4