Politis Lab
@Politis_Lab
Followers
447
Following
31
Media
20
Statuses
98
Structural Mass Spectrometry group at The University of Manchester lead by Argyris Politis. Tweets by lab members. #HDXMS #membraneprotein #structuralbiology
Manchester, England
Joined March 2017
We're hiring! Join us at @FBMH_UoM to work on cutting-edge #HDXMS of membrane proteins in native lipid environments. Exciting science – apply now!.
jobs.manchester.ac.uk
0
1
2
RT @NatureComms: Researchers from @omasstx and @Politis_Lab use HDX-MS to gain insight into structural dynamics of β1AR upon ligand binding….
0
4
0
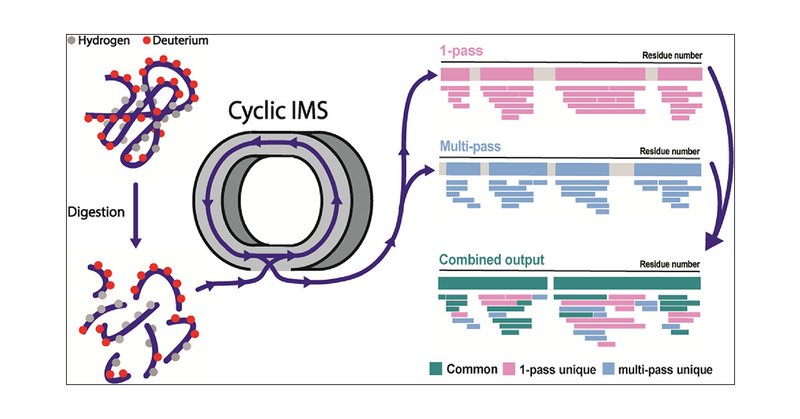
RT @dkwilson2: #HDX2024 Damon Griffiths is talking about how great cyclic ion mobility is for HDX-MS. Apparently, with multiple passes, it….
0
3
0
We have an exciting PhD CASE opportunity in our group in collaboration with OMass Therapeutics.
findaphd.com
PhD Project - (MRC DTP CASE) An Integrative Structural Biology Platform for Drug Discovery at The University of Manchester, listed on FindAPhD.com
0
1
2
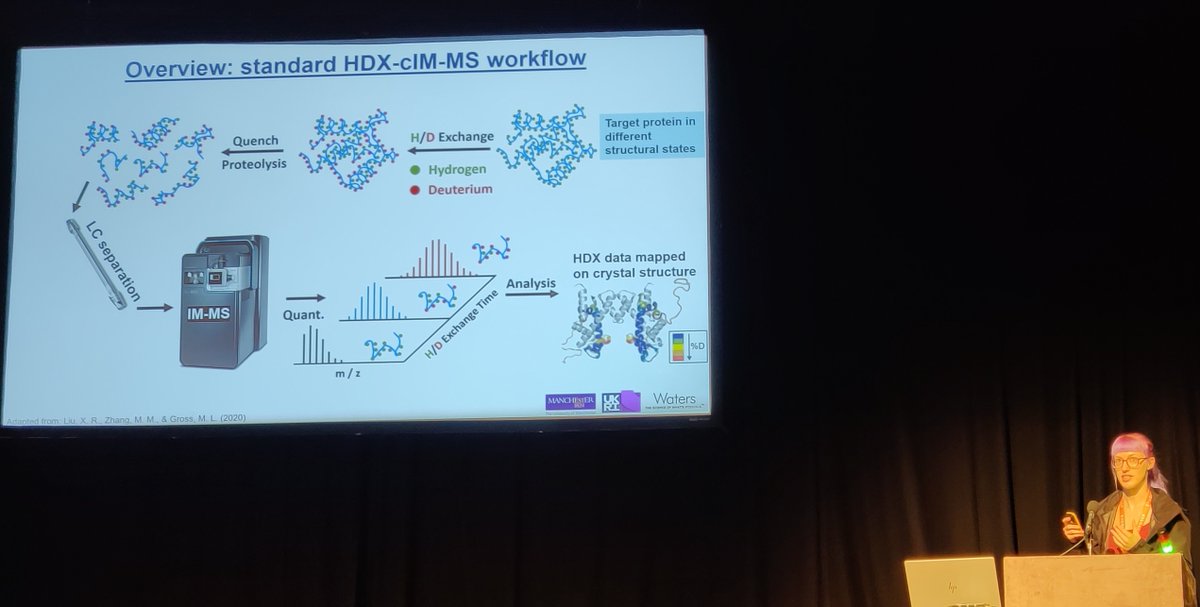
Check out our latest publication in @an_chem, where we integrate cyclic ion mobility (cIM) into HDX-MS workflows to enhance data quality and enable the analysis of more complex samples.
pubs.acs.org
Hydrogen/deuterium exchange-mass spectrometry (HDX-MS) has emerged as a powerful tool to probe protein dynamics. As a bottom-up technique, HDX-MS provides information at peptide-level resolution,...
0
2
6
We are looking for research technician in our group in Manchester. Please get in touch if interested:
jobs.manchester.ac.uk
0
1
2
We are excited to share this new preprint from our group, in which we utilised HDX-MS to uncover dynamic signatures of a class A GPCR. A great collaboration with OMass Therapeutics:
biorxiv.org
G-Protein Coupled Receptors (GPCRs) constitute the largest family of signalling proteins responsible for translating extracellular stimuli into intracellular functions. When dysregulated, GPCRs drive...
0
1
4
RT @kermitmurray: [ASAP] Hydrogen–Deuterium Exchange Mass Spectrometry with Integrated Size-Exclusion Chromatography for Analysis of Comple….
0
6
0
RT @HdxMs: Our next online seminar will certainly live up to expectation with FANTASTIC speakers David Schriemer and Elizabeth Komives (bot….
0
5
0
We're hiring! Come join us @kclchemistry for #HDXMS studies of membrane transporters in their native lipid environments. Closing 21-Apr-21 #ukchemjobs
0
22
16
RT @HdxMs: Alert! .Two more great speakers secured for our online seminar series. May 6th - David Schriemer (Uni. of Calgary).June 3rd -….
0
3
0
Congratulations to Doctor @KJHansen_ for completing a successful viva today! 🧑🎓The lab is proud of you and we are all looking forward to celebrate with you this important achievement! 🥳.
1
0
13
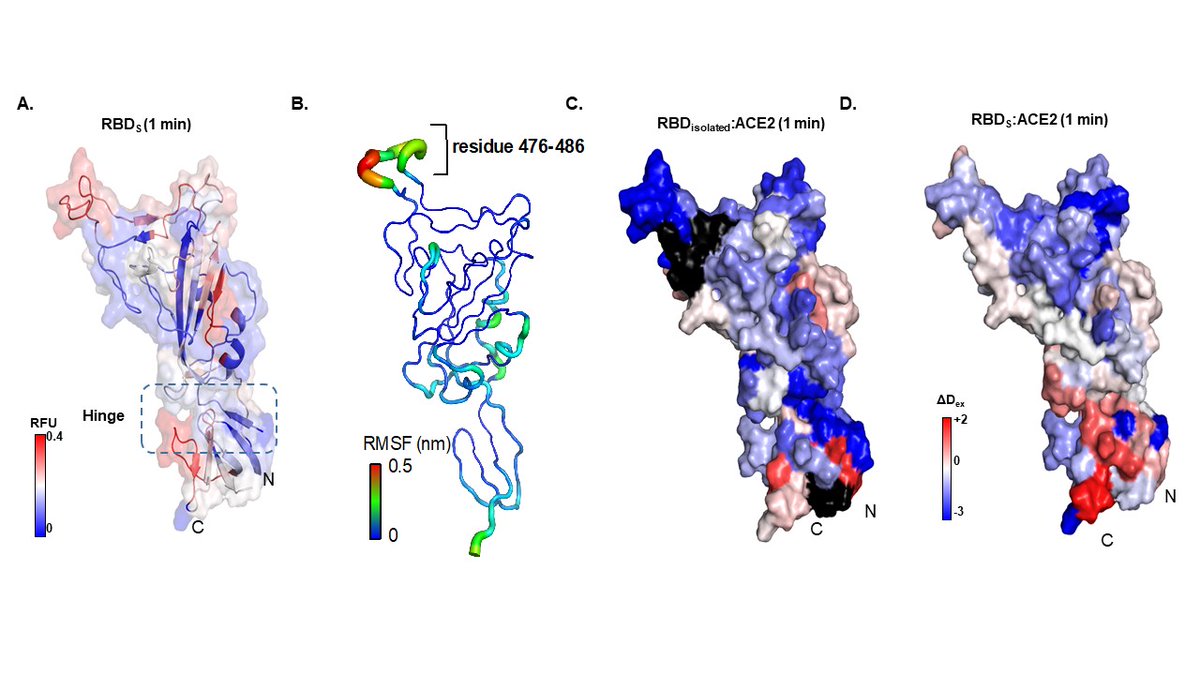
A very nice example on how HDX-MS is helping in one of the challenges of our century!.
Excited to share my group's (Thank you Raghu and Nikhil!) paper in eLife mapping Spike:ACE2 interface and allostery by HDXMS!. @eLife
0
0
3
Two exciting positions in the Department. Join us! (2) -->.
Reader in Computational Chemistry @kclchemistry . Salary: £57,631 - £64,405 p.a. Apply: Closes: 31 March 2021 . #KCLjobs #HiringNow #LondonJobs #JobAlerts #UniversityJobs.
0
0
1
Two exciting positions in the Department. Join us! (1) -->.
Lecturer in Chemistry @kclchemistry . Salary: £46,292 - £54,534 p.a. Apply: .Closes: 31 March 2021. #KCLjobs #HiringNow #LondonJobs #JobAlerts #UniversityJobs.
0
1
0
Read our newest publication in @NatureComms where we use HDX and MD to investigate the membrane transport protein XylE:
0
3
13
Wonderful collaboration with the Muller Group (@lab_mueller) at KCL. Check out the paper in Biochemistry:
0
5
19