Pacome Prompsy
@PPrompsy
Followers
41
Following
112
Media
1
Statuses
26
Bioinformatician, Data Engineer & Data Analyst at Institut Curie
Joined September 2020
Article published in NAR Genomics and Bioinformatics. Please try it out on , and post issues if encoutering any bugs. Happy to take any feedback !.
github.com
IDclust is an unsupervised method for clustering single-cell datasets (scRNA, scEpigenome). The cells are iteratively re-processed and re-clustered. Subclusters are created provided they are signif...
0
0
2
Pleased to present IDclust, a tool to automate and ease exploration of single-cell datasets. IDclust recursively clusters and find differential features between clusters until no more differences are found. With @MelissaSaichi @gama_search @VallotCeline
academic.oup.com
Abstract. The increasing diversity of single-cell datasets require systematic cell type characterization. Clustering is a critical step in single-cell anal
1
2
3
RT @VallotCeline: 📝Epigenomic heterogeneity as a source of tumour evolution. Just out in Nature Reviews Cancer - a piece we wrote with @Mat….
nature.com
Nature Reviews Cancer - Single-cell epigenomic technologies are refining our understanding of cancer evolution. Here Laisné et al. describe how epigenomic heterogeneity generates dynamic...
0
25
0
RT @VallotCeline: Our team is expanding, we are looking for a new collaborator - Computational Scientist (IR) - to join us at @institut_cur….
0
27
0
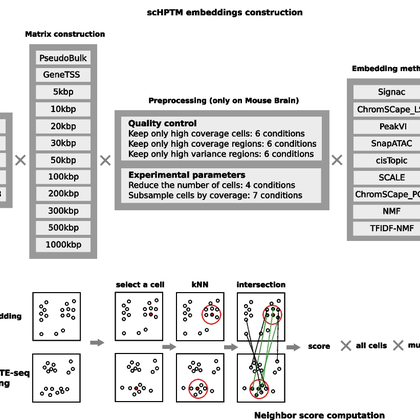
RT @gama_search: Our paper with @jeanphi_vert and @VallotCeline on how best to analyse single-cell histone modifications is finally out on….
genomebiology.biomedcentral.com
Background Single-cell histone post translational modification (scHPTM) assays such as scCUT&Tag or scChIP-seq allow single-cell mapping of diverse epigenomic landscapes within complex tissues and...
0
16
0
RT @VallotCeline: MAYA algorithm for scRNA is out in @NatureComms - it detects common expression programs across tumor cells from various c….
0
27
0
RT @gama_search: "Best practices for single-cell histone modification analysis" w/ @VallotCeline @jeanphi_vert and @PPrompsy. We identify….
0
10
0
RT @institut_curie: #Recrutement .Le Centre de recherche de l'Institut Curie recrute un #DataAnalyst/ingénieur de recherche (H/F) pour l'é….
0
7
0
RT @VallotCeline: Interested about epigenomics, want to learn how to analyze epigenomes at single-cell resolution? Join us!!.
0
27
0
RT @LabMorillon: The Morillon's group is recruting a 3 years post-doc to reveal what you never dare to ask on cancer dark transcriptome inv….
0
32
0
RT @mpmeers: Today there's a revised version of the MulTI-Tag manuscript out on bioRxiv--dig in here for a brief summary of some new improv….
biorxiv.org
Chromatin profiling at locus resolution uncovers gene regulatory features that define cell types and developmental trajectories, but it remains challenging to map and compare distinct chromatin-ass...
0
23
0
RT @VallotCeline: 1/5 Our manuscript on early breast tumorigenesis is out on biorxiv @institut_curie @INSB_CNRS,….
0
23
0
RT @VallotCeline: Merci la @lamethodeFC @NatachaTriou @NicoMartinFC pour cette mise en lumière de nos travaux sur le rôle des modifications….
0
5
0
RT @VallotCeline: Our study on the role of repressive histone mark H3K27me3 in determining cell fate upon chemotherapy treatment is out! Co….
0
42
0
RT @VallotCeline: Together with @MathildeJDura & @DeborahBourchis on the upcoming issue of Nature Genetics, all the way from spermatogenesi….
0
10
0
RT @NatureGenet: Out now! H3K27me3 conditions chemotolerance in triple-negative breast cancer (Marsolier et al.) ht….
0
15
0
RT @VallotCeline: We are looking for a talented postdoc to work on the evolution of epigenomes in the early steps of breast tumorigenesis i….
0
36
0
RT @VallotCeline: A new pre-print from our lab showing repressive histone landscapes are a determinant of cell fate under chemotherapy expo….
0
7
0