Weize Xu
@Nanguage
Followers
205
Following
598
Media
24
Statuses
64
Postdoc Scholar @Stanford BASE, Genetics | Build AI agent system for life science/biomedicine(PantheonOS: https://t.co/pdpUJ0jfMB)
Palo Alto, CA
Joined August 2015
All code datasets have been open-sourced👇: GitHub repo: https://t.co/S3nrNJgtM2 HuggingFace Dataset: https://t.co/wRhy18vPR2 Web app: https://t.co/7yCJHqOnsu Chatbot plugin: https://t.co/Oq66XEWsZ6 Napari:
napari-hub.org
Deep learning-based FISH spot calling method..
0
1
3
U-FISH has strong generalization capabilities: with minimal fine-tuning, it can be applied to signal spot detection tasks beyond FISH images, such as CISH spot detection and Hi-C loop detection. To make it easy to use, we developed a Napari plugin, a web application, and a
1
0
1
U-FISH was finally published in Genome Biology(@GenomeBiology )! 🥳U-FISH is a universal FISH, imaging-based spatial omics image spot detector. Trained on a rich dataset, it achieves SOTA-level accuracy and can work across different types of microscopy images without fine-tuning.
2
2
11
Exciting update on PantheonOS: Introducing Pantheon-Notebook & Pantheon-CLI — the first fully open-source, Python-based agentic tools that go beyond Claude Code in the field of data analysis. Pantheon-CLI runs entirely on your computer or server, supports 60+ tools and 50+
6
5
25
Ever imagined running Seurat with natural language and exploring your data step by step? No code This Pantheon-CLI case study shows a full single-cell workflow—normalization, clustering, QC, marker detection, and cell type mapping—all through conversation.
1
3
5
🎉Grateful shoutout to Zehua(@starlitnightly) for the incredible work on Pantheon-CLI and to Xiaojie(@Xiaojie_Qiu) for the steadfast support! Try Pantheon-CLI and share feedback—we’re building in the open. Next up: Pantheon-Lab (web) and our distributed agent framework,
🚀 Introducing PantheonOS ( https://t.co/NZM3wcHXbG): A Fully Open-Source Agent OS for Science PantheonOS began as a research project in my Stanford lab and has since evolved into a vision to redefine data science in the era of AI—starting with computational biology, especially
1
3
10
🚀 Introducing PantheonOS ( https://t.co/NZM3wcHXbG): A Fully Open-Source Agent OS for Science PantheonOS began as a research project in my Stanford lab and has since evolved into a vision to redefine data science in the era of AI—starting with computational biology, especially
2
38
116
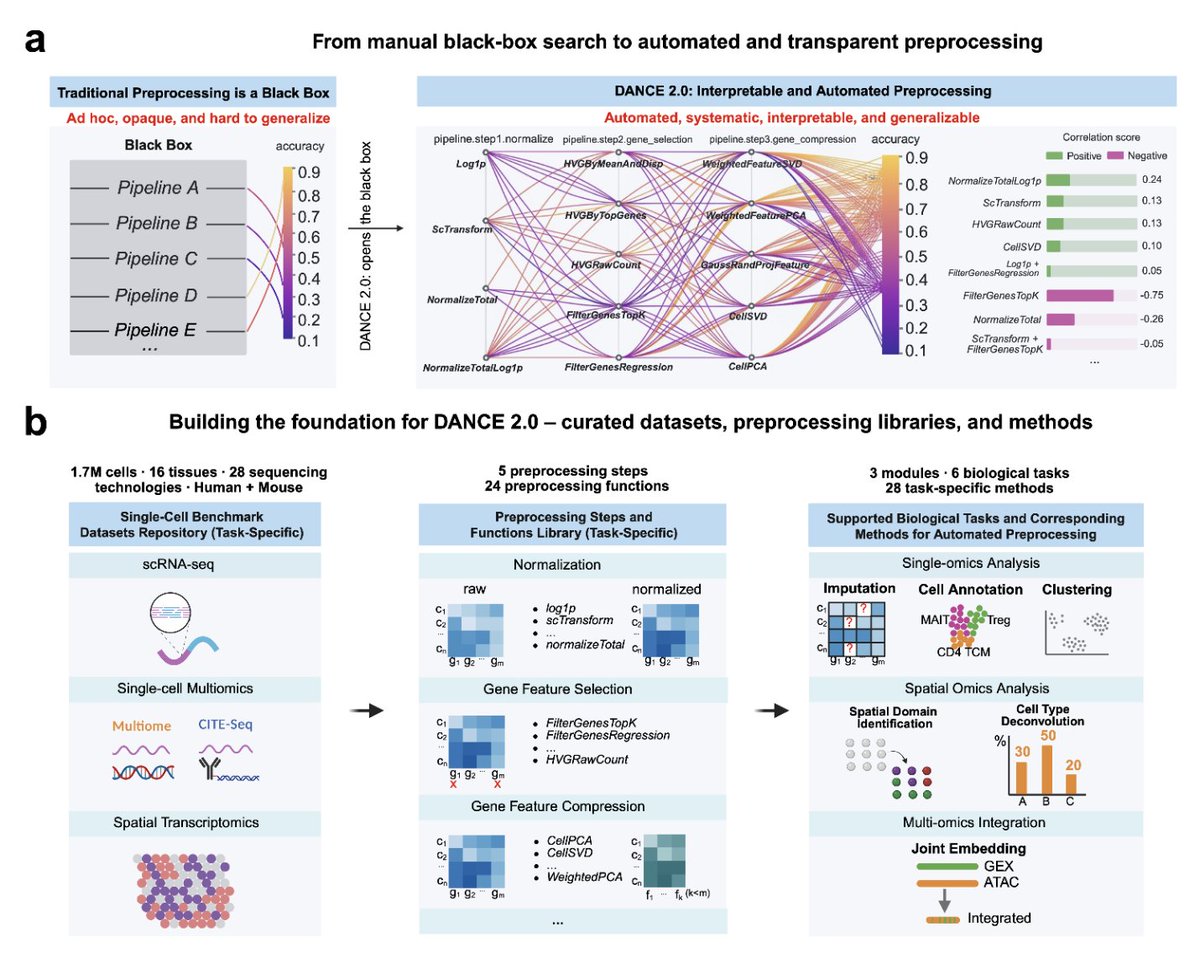
🚨 New to single-cell analysis? Still stuck in a trial-and-error loop? 🚨 Already experienced? Your “optimal” preprocessing pipeline might not actually be optimal. 🚨 Relying on a BioAgent? It likely runs standard procedures that are broadly accepted — but not truly optimal for
0
6
16
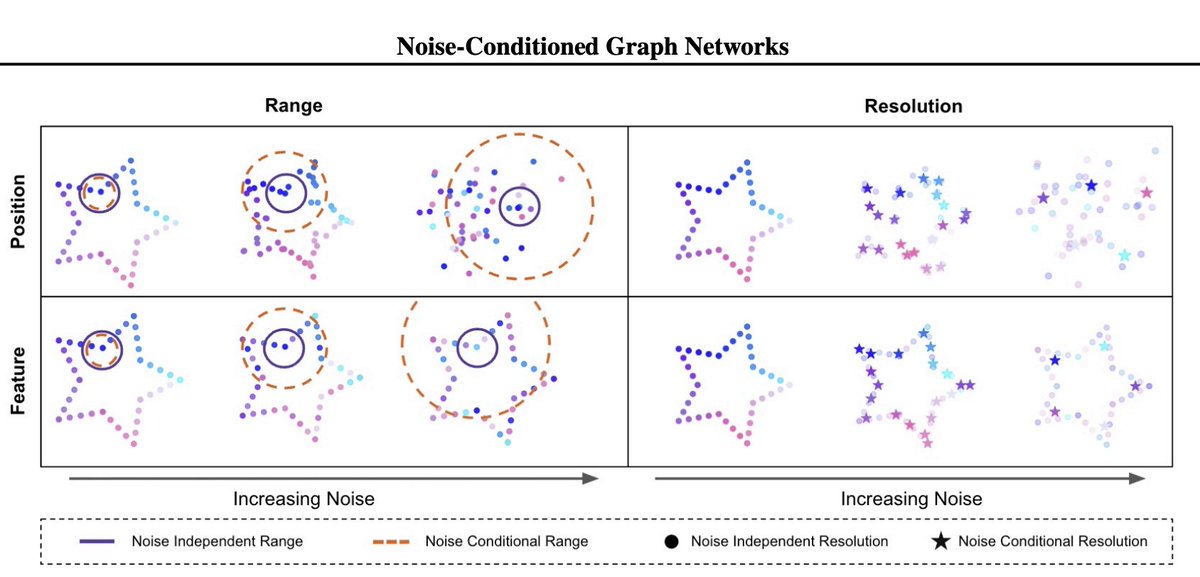
Why do diffusion models use the same GNN structure across denoising? Our #ICML paper presents Noise-Conditioned Graph Networks, a class of GNNs that adapts the graph structure to the noise level of the generative process. 📄 https://t.co/byfpZZ830O 💻 https://t.co/YKlJ5sacDs 🧵
1
6
27
I'm thrilled to share that a rotation project fully led by my first PhD student, the amazing Peter Pao-Huang @peterpaohuang, has been accepted to ICML 2025! Peter's paper, “Geometric Generative Modeling with Noise-Conditioned Graph Networks”, addresses a fundamental limitation
1
3
34
We are thrilled to share our new single-cell foundation model, Tabula (preprint: https://t.co/tkCi2l5KFi; package: https://t.co/bwkkgZBVnh)—a privacy-preserving predictive foundation model for single-cell transcriptomics, leveraging federated learning and tabular modeling. Over
1
28
147
It is a great honor to join the Qiu Lab(@Xiaojie_Qiu) at Stanford. Looking forward to contributing to valuable and interesting work in the future.
1
0
5
A different launcher API is also provided for quickly creating jobs, allowing you to rapidly set up a parallel data analysis workflow💪.
0
0
0
Moreover, you can exchange data with a generator in another process using the send method.
1
0
0
Executor engine supports generators, making it easy to create generators runing in another process.
1
0
0
🚀 Discover Executor Engine: A powerful, lightweight job management & parallel computing library for Python! ✨ Features: - Multiple job types (Thread, Process, Dask ...) - Easy parallel workflows - Flexible job conditions - Async/sync APIs https://t.co/ao3wloulB8
#Python
1
1
5
💻 If you missed the AI4Life's hackathon 2 weeks back, you can catch up on all the developments in this article: https://t.co/UmKjbNJrB1 Thanks to all the amazing participants and AI Health Innovation Cluster for the sponsorship!
0
12
22
Use SAM-like models in Fiji, finally.🥳🥳 I am pleased to introduce the early version of SAMJ, a plugin that enables semi-automatic annotation of microscopy images using Segment Anything Models. No GPU required(!!) Link to plugin page: https://t.co/Acv3xlGMMs
7
63
240
Happy to announce that our U-FISH preprint is now live 🎉!
U-FISH: a universal deep learning approach for accurate FISH spot detection across diverse datasets https://t.co/Yk9GPfxM3S
#biorxiv_bioinfo
0
2
14