MaizeGDB
@MaizeGDB
Followers
2K
Following
472
Media
269
Statuses
916
MaizeGDB is a community-oriented, long-term informatics service to researchers focused on the crop plant and model organism Zea mays.
the NET.
Joined February 2011
⚠️ Phishing alert: Someone is impersonating MaizeGDB and emailing links to download files from https://t.co/8mErkbntVn. These emails are NOT from us and may be malicious. MaizeGDB will never send unexpected download requests. Delete/report as spam or phishing if you receive one.
0
6
6
In Scientific Discoveries, learn how ARS researchers are using artificial intelligence (AI) to study fungal pathogens to help corn growers save their crops. Corn is the most important U.S. crop in terms of total production, with a value of $79.5 billion over the past four years.
0
8
17
New at MaizeGDB: The "Site Map" is a detailed launchpad to our tools, datasets, and resources. From the "Explore MaizeGDB" link in our menu, jump to Data Centers, JBrowse, BLAST, SNPversity, PanEffect, Phylostrata, Fusarium Protein Toolkit, and more. https://t.co/uzaakUIzbz
0
0
1
🌽New tool at MaizeGDB: Explore evolutionary origins of maize proteins! Search up to 100 genes to visualize conservation or to download homologs’ subcellular localization & GO terms! Whole-genome results available for download for B73 and the NAM founders. https://t.co/UND4KtXKGp
0
13
35
Newly published work from Elly Poretsky and @GrainGenes on understanding the structural variability of Pfam protein domains using AlphaFold2 across 16 model organisms. They observed substantial variation across Pfam families.
onlinelibrary.wiley.com
Understanding the biological functions of proteins is one of the main goals of functional genomics. Such understanding will help control and manipulate biological processes to enhance desirable...
0
1
4
microPublication (@Micropub7n) is a good resource to publish research from class or rotation projects. It is fast, peer‑reviewed, and citable. Here is a recent example from @DafangW. https://t.co/wQ6q7OgZpw
0
0
1
Why do some predicted protein structures fold poorly? Benchmarking AlphaFold, ESMFold, and Boltz in maize https://t.co/LqcCuwSfQW
#biorxiv_bioinfo
biorxiv.org
Protein structure prediction tools have significantly reduced the time and cost to generate protein structures and accelerated protein discovery and design. However, plant proteins are underreprese...
0
24
73
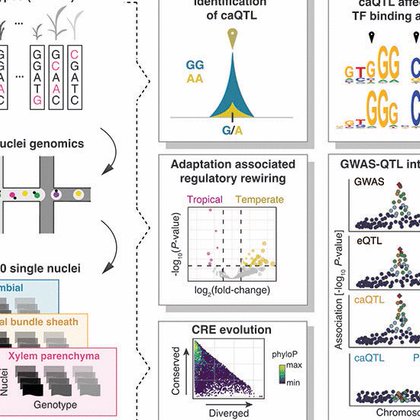
New data from the Bob Schmitz lab (Marand 2025) on cell type-specific cis-regulation in maize! This study, in @ScienceMagazine, reveals how DNA changes impact expression and maize traits. Explore the 176 tracks on our browser (Epigenetics and DNA Binding) https://t.co/4iZYlnAdn8
science.org
Gene expression and complex phenotypes are determined by the activity of cis-regulatory elements. However, an understanding of how extant genetic variants affect cis regulation remains limited. Here,...
1
16
44
We're excited to share PlantCAD browser tracks: a new, unpublished dataset integrating AI techniques for plant genome analysis. Explore it here: https://t.co/NasGqLw8Bt We welcome the community's feedback and innovative use cases for this data!
0
14
50
🌽 New browser tracks: Dr. Laura Tibbs-Cortes aligned proteomes spanning 11 evolutionary levels (from E. coli to sorghum) aligned to B73, NAM & PanAnd maize genomes! Additional tools coming soon! https://t.co/YfrnhjcM3H
0
2
4
Join us Thu, Mar 6 (2–5 pm) before the #MGM2025 for the Maize Genetics & Genomics Resources Workshop, "Shaping the Future of Maize Pan-Genomes." We'll have 8 talks on building & visualizing pan-genomes plus a discussion led by M. Hufford & C. Hirsch. https://t.co/j50mz5z9wJ
1
7
17
0
6
44
In addition, we have updated the web-based tool SNPVersity to explore the hundreds of millions of variant sites, integrating it with other tools at MaizeGDB.
0
1
4
The first release used a diverse set of 1,498 inbred lines, traditional varieties, and teosintes. Using a standardized variant-calling pipeline, we analyzed WGS data against the B73 reference genome. https://t.co/xBfjaQU3Ni
1
1
4
We need your input on which maize accessions to include in the next round of variant calling. In the upcoming 2025 release, we intend to remap ~2,500 new maize accessions. We aim to incorporate community feedback to make this resource more broadly useful. https://t.co/LklzZSQXRb
1
14
28
Congrats to Ed Buckler (USDA-ARS) who has been chosen as the 2025 recipient of The Barbara McClintock Prize for Plant Genetics and Genome Studies by the Maize Genetics Cooperation!
🎉Edward Buckler, plant geneticist w/ @USDA_ARS & CALS adjunct prof, has been awarded the 2025 Barbara McClintock Prize for Plant Genetics & Genome Studies from the Maize Genetics Cooperation, a global org of maize geneticists & breeders. Congratulations! https://t.co/SD7UOZqUbA
3
20
106
After a multi-year effort, a major SoyBase upgrade is nearly ready for release. The new SoyBase site is completely reworked, with new tools, & many interface improvements. Check out the NEW SoyBase & send your comments to the SoyBase team. https://t.co/YjZ6kEEtiL
1
6
18