Lea Lab
@LeaLabTweets
Followers
310
Following
24
Media
15
Statuses
101
Structural biology lab at NCI-Frederick. Formerly University of Oxford. Studying protein structures at the host-pathogen interface. All views are our own.
Frederick, MD
Joined February 2013
Thank you to @robinson_julia of Chemistry World (Royal Society of Chemistry) for a lovely write up of our work. Read the article here: And for the original paper:
1
6
30
RT @ChemistryWorld: Using cryo-electron microscopy two different groups have revealed new insights into how flagellum power bacteria to get….
0
7
0
RT @Dunn_School: New meningitis vaccine developed by the @TangLabOxford!. The new chimeric protein-based #vaccine to tackle #MenB is the re….
0
16
0
RT @NatureMicrobiol: Also hot off the press! Our latest article on the structural analysis of protein transport by the T9SS is out now 👇htt….
nature.com
Nature Microbiology - Cryo-EM analysis of the substrate-bound T9SS from Flavobacterium johnsoniae reveals an extended translocon complex and provides insight into protein secretion.
0
28
0
RT @kyliejwalters: Thrilled to share that we teamed up across @theNCI and @nih_nhlbi to discover #degraders against #hRpn13 that trigger #a….
link.springer.com
Nature Communications - Here, the authors identify a small molecule degrader (XL44) for hRpn13 and solve the XL44-hRpn13 structure. XL44 induces apoptosis in myeloma cells with hRpn13 dependency...
0
11
0
RT @S_M_Lea: Published today @SJohnsonDPhil and @justindeme85 story finally revealing the molecular basis for transmission of uni-direction….
0
46
0
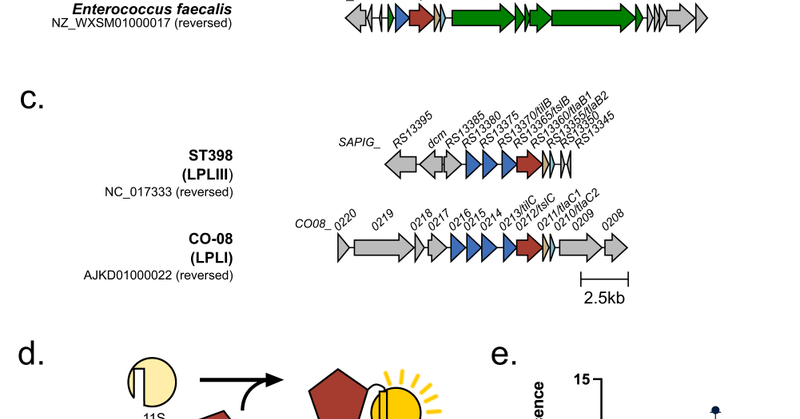
RT @OJ_Bryant: Check out our latest paper where we identify a new C-terminal export signal in flagellar Type III secretion substrates which….
journals.asm.org
Many bacterial pathogens utilize T3SS to inject virulence proteins (effectors) into host cells or to assemble flagella on the bacterial cell surface. Bacterial flagella present a paradigm for how...
0
4
0
RT @StephenRG97: Very happy to share some work from my PhD which is now published in Nature Communications! We have identified a new type o….
nature.com
Nature Communications - Here Garrett et al. describe a toxin, TslA, secreted by type VII secretion system that has a reverse domain arrangement compared to other previously characterised...
0
42
0
RT @OJ_Bryant: Check out our new paper @ from my time working in the fantastic Chung lab @TranslatingRNA. Also chec….
0
8
0
Fantastic opportunity to join a vibrant new lab at a great institute (we may be biased).
💥We're hiring! Two fully-funded postdoc positions at my lab at the NIH intramural program in Frederick, MD. We are mapping the structure of whole proteomes using crosslinking MS and structure modeling. At one of the most dynamic scientific institutes! Links are below.
0
0
0
And one more time to bacterial motility, with the companion paper to the gliding motility paper. Here are cryoEM structures of the flagellar stator complexes that drive rotation of the flagellum.
nature.com
Nature Microbiology - Using single-particle analysis cryogenic electron microscopy, the authors determine the structures of the bacterial flagellum stator complexes from three diverse bacteria.
1
0
1
We also have nonameric cryo-EM structures of flagellar and non-flagellar type III secretion system export apparatus proteins.
biorxiv.org
Type three secretion is the mechanism of protein secretion found in bacterial flagella and injectisomes. At its centre is the export apparatus (EA), a complex of five membrane proteins through which...
1
0
0
Another old lab favourite, the Notch signalling pathway. This pre-print characterises the Drosophila Notch and Delta/Serrate system.
biorxiv.org
Accurate Notch signalling is critical for organism development and homeostasis. Fine-tuning of Notch-ligand interactions have substantial impact on signalling-outputs. Recent structural studies...
1
0
1
And once more to bacterial motility, this time the structure of a motor that drives gliding motility.
nature.com
Nature Microbiology - Using cryo-electron microscopy, the authors describe the structure and function of a molecular motor powering both the Bacteroidetes type 9 protein secretion system and the...
1
0
1
Back to complement (and related serum cascades), and a cryoEM structure of C1-inhibitor bound to a Bordetella pertussis autotransporter protein.
journals.asm.org
The structure of a 10-kDa protein complex is one of the smallest to be determined using cryo-electron microscopy at high resolution. The structure reveals that C1-INH is sequestered in an inactivated...
1
0
2
And an old lab favourite, the complement system. This pre-print presents structures of a family of tick derived properdin inhibitors, including one bound to properdin.
biorxiv.org
Activation of the serum-resident complement system begins a cascade that leads to activation of membrane-resident complement receptors on immune cells, thus coordinating serum and cellular immune...
1
0
2