The Kinase Library
@KinaseLibrary
Followers
68
Following
5
Media
0
Statuses
10
Official account of the Kinase Library project: https://t.co/pEFtM3N1C6 All ideas shared on this account are free to use by anyone! #OpenScience
Joined January 2023
Finally out!.
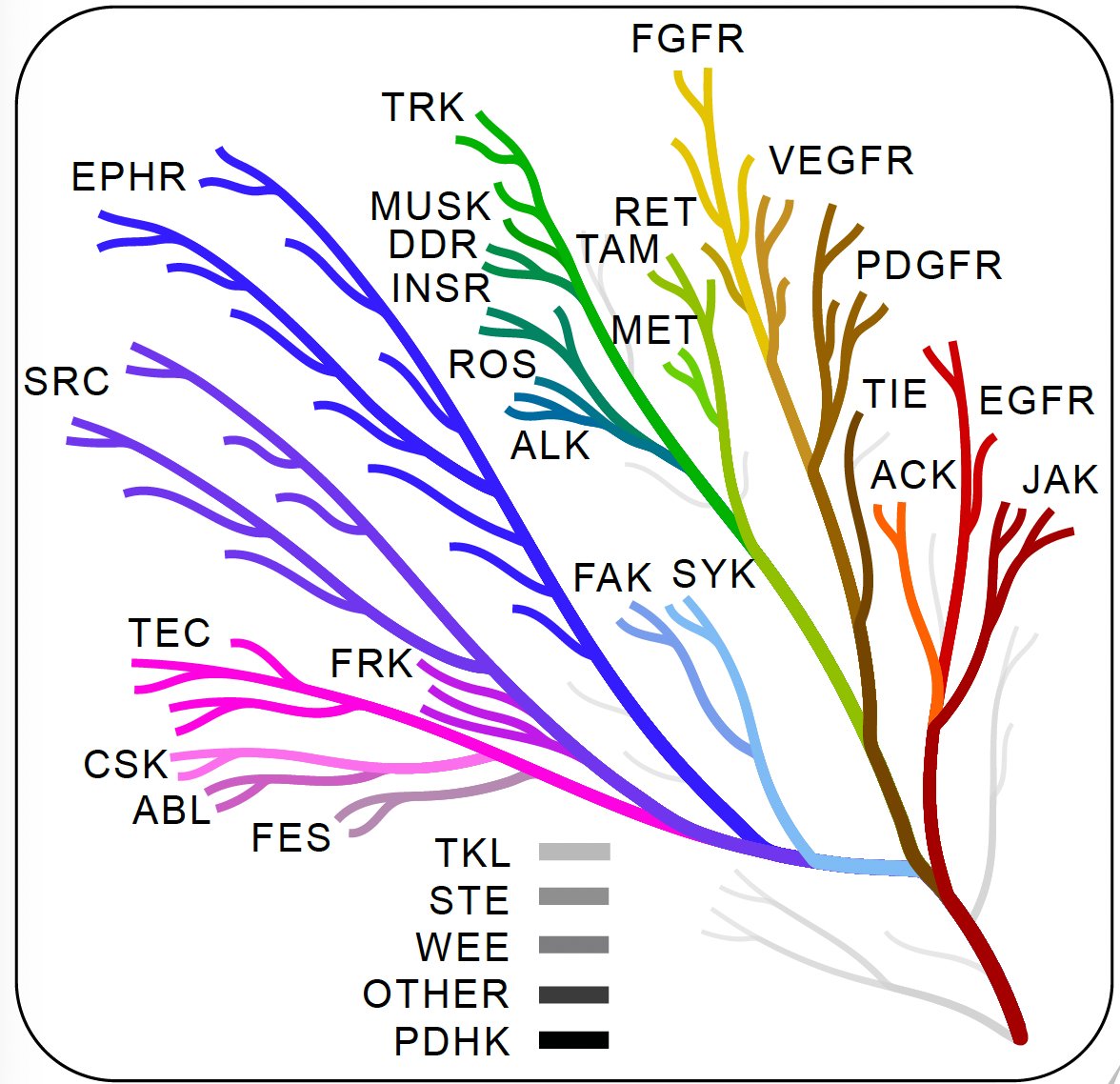
We are delighted to share the #tyrosine @KinaseLibrary, now in @Nature: Tyrosine predictions and enrichment analysis are now available: #TheKinaseLibrary #KL #part2. 🧵. 1/9.
0
0
4
Check out our new website!.
🚀 We just launched the brand-new #KinaseLibrary website, directly connected to the recently released Python package! Explore kinase-substrate relationships, run enrichment analyses, and visualize phosphoproteomics data—no coding required!. @KinaseLibrary.
0
0
1
🚀 TL;DR: pip install kinase-library 🚀. We are excited to introduce #TheKinaseLibrary Python package! 🧵👇. @KinaseLibrary Jared_Johnson @Tomer_M_Yaron @CellSignal.
0
0
1
RT @Tomer_M_Yaron: The tyrosine kinome was my very first rotation project back in October 2017. I used to take the entire time during lab m….
0
3
0
RT @Nature: Nature research paper: The intrinsic substrate specificity of the human tyrosine kinome
0
31
0
RT @CantleyLab: We are delighted to share our work, now available in .@Nature:. Our scoring system can be accessed….
0
305
0
RT @CantleyLab: We are proud to introduce our global atlas of substrate specificities for the human serine/threonine kinome, now on bioRxiv….
0
124
0