Justin Lemkul
@JustinLemkulVT
Followers
3K
Following
2K
Media
235
Statuses
5K
Associate Professor of Biochemistry at Virginia Tech. Drude FF contributor, #compchem, simulations, and CADD. Tweets are my own. RT != endorsements. Go #Hokies!
Blacksburg, VA
Joined February 2014
Come be my colleague (pt. 3, yes it's true)! Another non-TT instructional faculty position in our brand new Discovery Space! An amazing opportunity for hands-on, experiential learning with undergrads. #chemjobs #facultychemjobs @Chemjobber
https://t.co/BhmHP3Gs6U
0
2
6
Come be my colleague (pt. 2)! We have an opening for a Collegiate Assistant Professor (non-tenure track instructional faculty) to teach and innovate in our senior-level laboratory class. Join us! #chemjobs #facultychemjobs CC: @Chemjobber
https://t.co/mUwaceMh33
1
6
6
Come be my colleague! The Department of Biochemistry at Virginia Tech is hiring a tenure-track assistant professor in the area of macromolecular complexes. Please see the job ad linked below. #chemjobs #biochemjobs #facultychemjobs CC: @Chemjobber
https://t.co/k2Gnc4CneF
0
12
19
Adv multiscale simulation software predicting protein-ligand (un)binding kinetics for real systems SEEKR now powered by #metadynamics (+ QM + MD + BD + milestoning) k_on, k_off & deltaG for thr-tyr kinase Collab w/ @SHIONOGI_JP @a_anand_ojha
https://t.co/dVoD2yU6rm
1
37
208
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
514
9K
20K
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Physics to John J. Hopfield and Geoffrey E. Hinton “for foundational discoveries and inventions that enable machine learning with artificial neural networks.”
1K
13K
33K
BREAKING NEWS The 2024 #NobelPrize in Physiology or Medicine has been awarded to Victor Ambros and Gary Ruvkun for the discovery of microRNA and its role in post-transcriptional gene regulation.
545
11K
28K
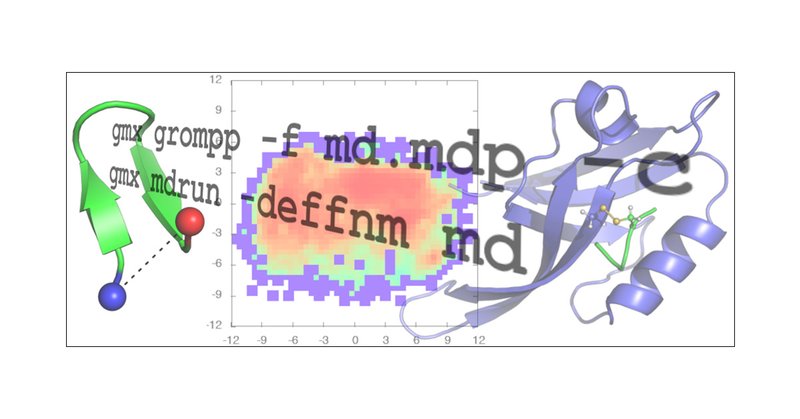
This article from @JustinLemkulVT of @vtbiochem provides updated tutorials for the GROMACS software. Users are presented with several common applications in simulating protein dynamics, with an emphasis on robustness and cautions against common pitfalls. https://t.co/PSsZya1gn7
pubs.acs.org
Atomistic molecular dynamics (MD) simulations have become an indispensable tool for investigating the structure, dynamics, and energetics of biomolecules. Continual optimization of software algorit...
0
23
69
If it takes over 5 minutes to reverse a call, it is not clear and indisputable.
0
2
12
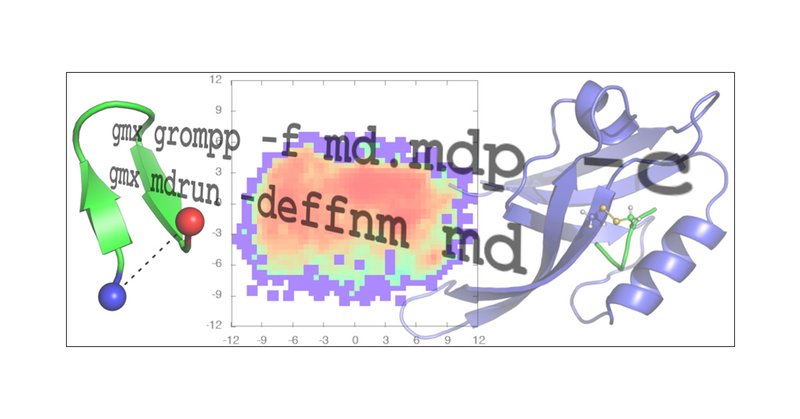
And I have a second one - updated tutorials for @GMX_TWEET version 2024! These are new and distinct from the other set of tutorials published a few years ago (those will get a refresh, too). I hope they are useful! #compchem #OA #GROMACS
https://t.co/T3tazfn7Zi
pubs.acs.org
Atomistic molecular dynamics (MD) simulations have become an indispensable tool for investigating the structure, dynamics, and energetics of biomolecules. Continual optimization of software algorit...
0
33
85
New articles out this week in @JPhysChem! This article becomes is the new citation for the current versions of the CHARMM program. What an incredible team of developers to be a part of. #compchem #OA
https://t.co/Evrkm7WNPi
1
13
43
One simulation is one observation. You wouldn't run an enzyme assay one time and claim an error bar of +/- 0. Same with simulations. Run replicates - different starting velocities, different initial configurations, etc. And plan your analysis! Again, don't just repeat tutorials.
0
1
14
Length of the simulation and assessment of convergence are CRUCIAL. Just because a tutorial does 100 ns does NOT mean that is enough time for your application. Maybe it is, maybe not. You have to prove it. You also have to run replicates. Full stop.
1
2
10
As someone who writes a lot of tutorial material (new ones just accepted for publication, stay tuned!), I see my protocols parroted back in lots of papers. Tutorials are highly idealized systems, for didactic purposes. When doing "real" science, you have many decisions to make.
1
1
8
Thank you to @spy_sci for the kind words; I will emphasize everything said in this thread. Anyone doing MD or thinking about it - please read. So many common problems that I have seen as a reviewer or editor. Computational work requires deep expertise, it's not an "add-on."
Came across this open advertisement for collaboration on one of the bioinformatics and molecular modeling groups on facebook. This felt so wrong to me. If you want to do Molecular Dynamics Simulations but do not have expertise or knowledge then this 🧵 is for you.
1
16
62
New @ChemRxiv preprint up from @mdpoleto on the parametrization of a polarizable model for PET polymer. Really interesting induced polarization effects in this material and potential interactions with proteins! #compchem
https://t.co/Dif5IFpqIO
0
5
18
Today, we officially say adeus and boa sorte to @mdpoleto, who was an outstanding postdoc in our group for ~4 years. Thank you for all your contributions and best wishes as you start your independent career at the University of São Paolo!
1
1
51
#ACSSpring2024: "we are sending scheduling info in the wrong time zone." #ACSFall2024: "we are sending badges to totally random people who aren't even attending."
0
0
11