John Pulice
@JohnPulice
Followers
595
Following
46K
Media
78
Statuses
1K
Scientist at @foghorntx | PhD @BBS_Harvard in the @meyersonlab | Previously: @kadochlab @harvard ‘15 @TJHSST ‘11
Cambridge, Massachusetts
Joined June 2011
Excited to share the final published version of my PhD work out today in @MolecularCell! See below for a thread of updates for the final version from the preprint! 1/
cell.com
Pulice and Meyerson uncover the activation and regulation of NKX2-1, the most significantly amplified gene in lung adenocarcinoma. Focal super-enhancer amplification drives NKX2-1 activation in LUAD....
2
2
11
Cool new paper from the @WJGreenleaf lab on TF dosage by ATAC-seq!
biorxiv.org
Transcription factor (TF) dosage is a critical determinant of cellular identity. However, the quantitative relationship between TF dosage and its regulation of chromatin accessibility and gene...
1
0
0
And if you're interested, check out our study on how NKX2-1 functions as a pioneer-like factor to drive lineage addiction through enhancer remodeling in lung adenocarcinoma!
cell.com
Pulice and Meyerson uncover the activation and regulation of NKX2-1, the most significantly amplified gene in lung adenocarcinoma. Focal super-enhancer amplification drives NKX2-1 activation in LUAD....
0
0
2
RT @manorlaboratory: My lab’s 5-year NIH R01 grant, awarded to study gene therapy for hearing loss, was abruptly terminated. I want to shar….
0
2K
0
RT @MolecularCell: Amplified dosage of the NKX2-1 lineage transcription factor controls its oncogenic role in lung adenocarcinoma https://t….
0
7
0
RT @MolecularCell: Online Now: Amplified dosage of the NKX2-1 lineage transcription factor controls its oncogenic role in lung adenocarcino….
0
20
0
@MolecularCell We've endeavored to make all the reagents and methods in this article as transparent and reproducible as possible. We deposited all plasmids (inc. pDONR cloning backbones and dTag OE destination vectors) on Addgene for easy use 15/.
0
0
0
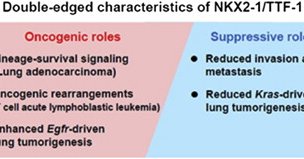
@MolecularCell NKX2-1 is a complicated gene in cancer, with conflicting data for its role as a tumor suppressor and oncogene. Our data suggest NKX2-1 is a common oncogene and rare tumor suppressor in human LUAD, with it's TS role restricted to the rare IMA subtype 14/.
cell.com
Emerging evidence indicates that NKX2-1, a homeobox-containing transcription factor also known as TTF-1, plays a role as a “lineage-survival” oncogene in lung adenocarcinomas. In T cell acute...
1
0
0
@MolecularCell Overall, we find NKX2-1 is a central lineage-defining oncogene in LUAD, and map both the activation and regulation of a critical oncogenic TF. The role of NKX2-1 in LUAD has been conflicted—we hope this clarifies a widespread role for NKX2-1 as an oncogene in human LUAD 13/
1
0
0
@MolecularCell Wanted to highlight a few experiments that are in the supplement:.(1) Dose-dependent proliferation and CDKN2B as a biomarker of cell cycle arrest. (2) Mapping functional domains of NKX2-1 in EGFR TKI persistence.(3) The effect of NKX2-1 OE/KD is specific to EGFR TKIs .12/
1
0
0
@MolecularCell For our interrogation of EGFR TKI persistence, we added new CTG, cell proliferation, and competition assay data to better understand this effect. We find that NKX2-1 drives an increase in cell survival and a decrease in maximum cell death from EGFR TKIs 11/
1
0
0
@MolecularCell We expanded our interrogation of NKX2-1 dependency in cell lines, finding our dependency effects extend to 3D colony formation and lead to cell cycle arrest. 10/
1
0
0
@MolecularCell Conversely, we found that NKX2-1 knockdown had predominantly linear effects, with even small decreases in TF dosage by shRNA leading to significant changes in gene expression and chromatin accessibility. 9/
1
0
0
@MolecularCell We have markedly expanded our interrogation of dosage modulation in both OE and KD settings. We find both thresholded and linear effects of NKX2-1 OE, with many genes and sites only regulated by EF1a::NKX2-1 but unable to be regulated by the intermediate SFFV::NKX2-1 level 8/
1
0
0
@MolecularCell We also used nascent RNA profiling by TT-seq to identify the direct targets of NKX2-1 regulation in this degron system 7/
1
0
0
@MolecularCell Using a degron system, we both overexpressed and degraded NKX2-1, and identified direct regulation of chromatin accessibility genome-wide. Sites opened by NKX2-1 OE were closed by 2hr of NKX2-1 degradation. 6/
1
0
0
@MolecularCell We have marked expanded the mechanistic interrogation of NKX2-1 regulation of chromatin accessibility and gene regulation in LUAD, using both degron systems to identify direct targets, and dosage modulation by overexpression and knockdown to characterize the dosage dynamics 5/.
1
0
0