David Emms
@David__Emms
Followers
598
Following
96
Media
19
Statuses
147
Developer of OrthoFinder and https://t.co/pWdIdp8atF, interested in phylogenomics and all things trees. Former postdoc in the @Steve__Kelly lab.
Joined September 2017
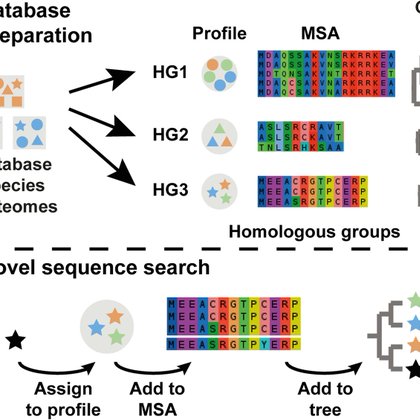
SHOOT, the phylogenetic sequence search engine, has been published!. SHOOT is like BLAST, but instead of a list of similar sequences you get a maximum likelihood phylogenetic tree for your query gene sequence. Try it👉 Article👉
genomebiology.biomedcentral.com
Determining the evolutionary relationships between genes is fundamental to comparative biological research. Here, we present SHOOT. SHOOT searches a user query sequence against a database of phylog...
19
469
2K
RT @dr_belcher: OrthoFinder just dropped a major update. It’s faster, more accurate, and ready for thousands of genomes. Let’s break it dow….
biorxiv.org
Here, we present a major advance of the OrthoFinder method. This extends OrthoFinder’s high accuracy comparative genomic framework to provide substantially enhanced scalability and accuracy. Specif...
0
11
0
RT @Steve__Kelly: It has been a while… but an updated, faster & more accurate version of OrthoFinder is now out! . Scales to thousands of….
biorxiv.org
Here, we present a major advance of the OrthoFinder method. This extends OrthoFinder’s high accuracy comparative genomic framework to provide substantially enhanced scalability and accuracy. Specif...
0
62
0
I've moved to new pastures, working on AI models for genomics at the excellent @instadeepai, but will carry on supporting the OrthoFinder team in the background. They're going to be working on some great, new functionality so keep an eye open for that: .
github.com
Phylogenetic orthology inference for comparative genomics - OrthoFinder/OrthoFinder
0
0
5
And with the new OrthoFinder team we have a new website to bookmark for all the latest updates:
github.com
Phylogenetic orthology inference for comparative genomics - OrthoFinder/OrthoFinder
1
0
3
We have a new version of OrthoFinder:. Faster scaling to larger species sets, phylogenetically resolved orthogroups, reduced RAM usage. Thanks to @dr_belcher, Yi Liu & Jonathan Holmes who have joined the team, and as ever OrthoFinder's PI @Steve__Kelly!.
biorxiv.org
Here, we present a major advance of the OrthoFinder method. This extends OrthoFinder’s high accuracy comparative genomic framework to provide substantially enhanced scalability and accuracy. Specif...
1
12
36
RT @Gloveface: We are looking for a postdoc in comparative genomics/bioinformatics! They will be working in a collaborative environment on….
0
125
0
RT @Steve__Kelly: Please RT . Last few days left to apply! . 4 positions to join us at Oxford, driving the next phase of development to Ort….
0
70
0
RT @GenomeBiology: SHOOT, from @David__Emms & @Steve__Kelly, searches for a sequence in a database of phylogenetic trees, identifies the ho….
0
15
0
is powered by OrthoFinder, DIAMOND, IQ-TREE, MAFFT, EPA-ng & phylotree.js with some other clever technologies under the hood to accelerate your search and gene tree inference. Try it👉 Article👉
genomebiology.biomedcentral.com
Determining the evolutionary relationships between genes is fundamental to comparative biological research. Here, we present SHOOT. SHOOT searches a user query sequence against a database of phylog...
2
2
25
You give a query sequence and it builds a gene tree and identifies the orthologs of your gene in other species. Try it👉 Article👉
genomebiology.biomedcentral.com
Determining the evolutionary relationships between genes is fundamental to comparative biological research. Here, we present SHOOT. SHOOT searches a user query sequence against a database of phylog...
2
1
20
RT @moodytomato: We have TWO fully funded PhD studentships available in our group to study 3D growth in plants. Both studentships are avail….
0
206
0
RT @_johntlovell: The preprint describing our comparative genomics #Rstats package is on bioRxiv (link below). GENESPACE uses synteny to ex….
0
208
0
RT @PlantTeaching: Plant Science Research Weekly .X-species sRNAs, SynBio -> N fixation, gene evolution & organelle….
0
11
0
RT @_johntlovell: Our GENESPACE comparative genomics R package is on github! GENESPACE conducts synteny-constrained comparisons of gene cop….
0
79
0
The source code for the command line version is on GitHub: There's also code on GitHub for a SHOOT webserver. If you're interested in adapting it to enable phylogenetic gene search for a genome resource website, let me know!.
github.com
SHOOT.bio - the phylogenetic search engine. Contribute to davidemms/SHOOT development by creating an account on GitHub.
0
1
2
The preprint for the gene search website with @Steve__Kelly has been updated: SHOOT lets you search a protein sequence & get back a maximum likelihood gene tree for your sequence. It's a robust & easy way to get accurate orthologs.
2
15
54
RT @Physacourses: Registrations are now open for the 4th edition of the #ComparativeGenomics course with @sedlazeck and Ingo Ebersberger in….
0
10
0