DassanayakeLab
@DassanayakeLab
Followers
107
Following
2
Media
7
Statuses
20
Joined September 2018
Check the newest pre-print from @DassanayakeLab and collaborators for comparative #transcriptomics of using #singlecellRNAseq on Diversification of gene expression across extremophytes and stress-sensitive species in the Brassicaceae https://t.co/H0uncCe8jM
biorxiv.org
Stress-sensitive and stress-adapted plants respond differently to environmental stresses. To explore the cellular-level stress adaptations, we built root single-cell transcriptome atlases for diverse...
0
3
7
We are hiring a postdoctoral researcher to work with invasive species genomes and designing targeted genome editing to control aquatic and terestrial invasive plants. Job description: https://t.co/BEpARkH5RF Apply here: https://t.co/yV0n0EhtVK Please share within your networks.
0
5
8
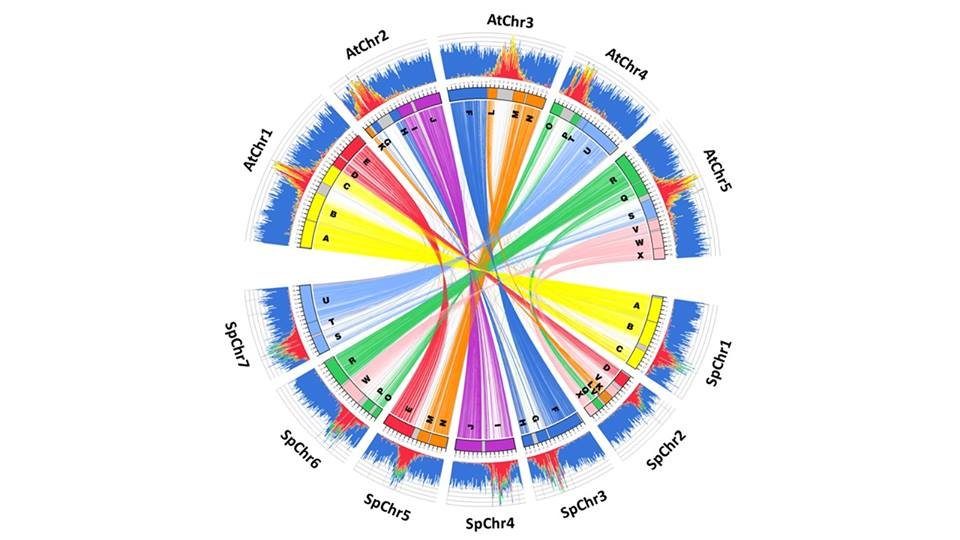
Congratulations Chathura for the Ph.D. defense. His most recent work can be read below. 1. Transcriptome atlas of Schrenkiella parvula; https://t.co/5dSrKWmYr5 2. Alternative splicing of Schrenkiella parvula under salt stress; https://t.co/rjhwzlu2Um
0
1
11
Good job @KieuNgaTran3 Multiple paths lead to salt tolerance - pre-adaptation vs dynamic responses from two closely related extremophytes Link for the paper: https://t.co/JZ2AdR2fNx
2
2
13
Welcome Thu and Samadhi to The Dassanayake lab #fall2021 #graduateschool #lsuresearch #lsubiologicalsciences
0
0
10
Balancing growth amidst salinity stress – lifestyle perspectives from the extremophyte model Schrenkiella parvula Link for the paper: https://t.co/s0WhgymhEU
0
0
4
Good job! @Pramodpanth
A tale of two transcriptomic responses in agricultural pests via host defenses and viral replication https://t.co/WYtaIRTzfq
#bioRxiv
0
0
6
Guannan Wang from our lab is presenting his work on plant adaptations to high Boron stress (6:20 pm; Extremophile Genomics; W402; Pacific Salon2) at the 2020 Plant and Animal Genome meeting, San Diego. @LSUResearch @LSU_BioSci #PAG2020
0
1
8
PhD in Genomics We are looking for enthusiastic graduate students to join our lab in 2020. They will be invited to join two recently funded NSF & DOE projects in the lab. Email maheshid@lsu.edu for details. NSF project: https://t.co/9w3JlGXuqp DOE project:
0
9
10
Our lab featured on LSU College of Science news. Where Will Our Farmlands be in the Next Decade? https://t.co/C6IME0syON We are looking for a new graduate student to join our lab in 2020 Fall. Please RT.
0
2
7
We were recently awarded an NSF-EDGE award to develop functional genomics tools for our extremophyte models, Schrenkiella parvula and Eutrema salsugenium. Link to the abstract;
0
1
7
We were recently awarded an NSF-EDGE award to develop functional genomics tools for our extremophyte models, Schrenkiella parvula and Eutrema salsugenium. Link to the abstract;
0
1
7
We are presenting our work at #LPSC2019 on growing plants on Mars. New experience with a lot of good discussion about habitation on Mars and other planets. A new area to discover more for plant biologists. #LSU_BioSci #LSUResearch
0
0
3
Genome browsers for extremophyte models, Schrenkiella parvula and Eutrema salsugineum are now available. Please check it out in the following link; https://t.co/xFn2Lzjbgv
@LSU_BioSci
0
2
2
The Agrobacterium-mediated floral dip transformation for the extremophyte Schrenkiella parvula is now published. The full paper and the video is available in the following link;
0
2
3
It was nice to have Dr. Simon Barak from Jacob Blaustein Institutes for Desert Research Ben-Gurion University of the Negev, Israel as a CDIB/BMB seminar speaker at LSU. In collaboration with his group, we have recently published a review article; Link; https://t.co/TN37J8cqFb.
0
4
7
We have recently developed the CLfinder-OrthNet pipeline. This is useful to detect co-linearity among multiple closely related genomes, finds orthologous gene groups, and encodes the evolutionary history of each orthologue group into a network (OrthNet). https://t.co/iDt6puq5k1
academic.oup.com
Abstract. We developed the CLfinder-OrthNet pipeline that detects co-linearity among multiple closely related genomes, finds orthologous gene groups, and e
0
1
1

