DavidMacPherson
@DMacPhersonLab
Followers
357
Following
90
Media
29
Statuses
109
Cancer Biologist @fredhutch
Seattle, WA
Joined February 2019
📣The MacPherson lab @fredhutch is hiring a postdoc to study the biology of small cell lung cancer. We employ in vivo models, genetic screens and genomic approaches to understand + therapeutically target subsets of SCLC. $78K salary ⬆️ to >$90K after 2 yrs https://t.co/pB0DbKd9ct
2
11
20
The Division of Human Biology @fredhutch is recruiting for a lab-based scientist at the Assistant or Associate Professor level. Come join a dynamic research community committed to pushing the boundaries of discovery in cancer biology. https://t.co/jAnG2L3YHJ
1
11
37
Thanks for highlighting our efforts to use in vivo CRISPR screens to identify drivers of resistance to chemotherapy in small cell lung cancer
In work published in Science Advances, researchers in the @DMacPhersonLab used #CRISPRCas9 on #SCLC tumor cells to better model how tumors that initially respond to chemotherapy become resistant.
0
1
5
This work was possible only because of the contributions of our collaborators – Anish Thomas, @charlesrudin, Keith Eaton, McGarry Houghton, Caroline Dive, as well as SCLC patients and funding from NCI and @KuniFoundation 14/14
0
1
3
Our fragmentomics approach complements other emerging strategies towards SCLC liquid biopsy phenotyping such as DNA methylation analyses and ChIPseq approaches. We advocate for broad application of different phenotyping approaches to patient samples 13/X.
1
0
1
We are continuing to validate the performance of SCLCpheno-seq, identify additional informative sites, and incorporate new phenotyping modules into the targeted panel to comprehensively phenotype SCLC and better understand therapeutic responses 12/X
1
0
0
As targeted sequencing is also used to call driver gene mutations, we layered on mutation analyses to develop an all-in-one assay to also call exonic mutations in >800 cancer-relevant genes, along with TF activation 11/X
1
0
0
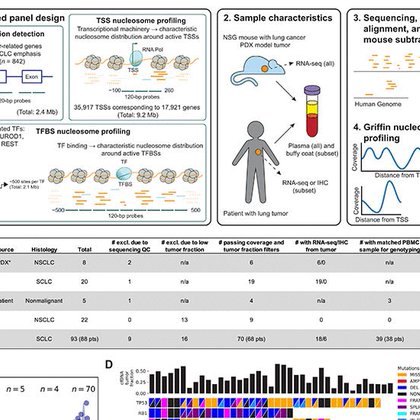
Probabilistic models focusing only on informative sites in PDXs distinguished NSCLC from SCLC in patients, suggesting utility in calling NSCLC-SCLC transformation. Our assay also showed strong performance at calling SCLC patient subsets for which TF expression was available 10/X
1
0
0
Many informative TSS sites from ctDNA included known lineage-defining TFs and their targets/collaborators. For example, POU2F3 TSS activity matched RNAseq expression and matched TSS activity of its co-activator C11orf53/POU2AF2 9/X
1
0
0
Joe then developed a clustering approach to distinguish TFBSs and TSSs where ctDNA coverage profiles synched with gene expression vs those that were non-informative. By analyzing PDX models, we could identify informative sites using an idealized system with 100% tumor ctDNA 8/X
1
0
0
From the ASCL1, NEUROD1, POU2F3 and REST TFBS regions, aggregating all sites together did well for some TFs, such as REST, NEUROD1 and POU2F3, but not for ASCL1 7/X
1
1
1
We built an assay to capture potentially informative regions including most genomic transcriptional start sites (TSSs) along with hundreds of TFBS regions for key TFs: ASCL1, NEUROD1, POU2F3 and REST. 6/X
1
0
0
Cell-free DNA is protected from nucleases by nucleosomes, while active regulatory regions such as TF binding sites and TSS regions get degraded. Deep WGS shows a nucleosome-depleted region at the TSS of NEUROD1 only in the NEUROD1-active SCLC patient/PDX model samples. 5/X
1
0
0
Also, EGFR-mutant non small cell lung cancer (NSCLC) can transform to SCLC as a mechanism of resistance to EGFR inhibitors, but the field lacks a liquid biopsy assay to detect such transformation 4/X
1
0
0
Activation of transcription factors ASCL1, NEUROD1, POU2F3, and repressor of neuronal/neuroendocrine state, REST defines distinct subsets of SCLC with differing therapeutic vulnerabilities. How can we distinguish TF activation in SCLC from blood samples? 3/X
1
0
0
Correlative analyses to reveal features of SCLC with differential therapeutic responses in clinical trials are limited by challenges in obtaining tissue biopsies. SCLC has remarkably high levels of ctDNA, leading to an opportunity to phenotype SCLC from liquid biopsies 2/X
1
0
0
Our collaborative study with @GavinHa @fredhutch is out in @ScienceAdvances. Led by Joe Hiatt and Anna Lisa Doebley, we developed a liquid biopsy approach, SCLCpheno-seq, to classify SCLC subsets and distinguish from NSCLC based on analyses of ctDNA 1/X
science.org
Targeted cell-free DNA nucleosome profiling at transcriptional regulatory regions can distinguish lung cancer types and subtypes.
1
3
16
We have an open joint-postdoc position with @DMacPhersonLab to work on new computational approaches for studying #lungcancer using #liquidbiopsy. If you're interested in cancer genomics, LBx, AI or #PrecisionOncology, pls apply! https://t.co/IN4uOWTRy0
0
5
9
I’m counting down the sleeps to @IASLC #SCLC23! Was thrilled to help organise this meeting together with @AlvaroQuintanal @DMacPhersonLab the twitter-less Kathryn Simpson & @charlesrudin. Don’t hesitate - register now! 👇
Join us for #SCLC23! A meeting focused on preclinical/clinical advances in #SCLC research, including basic research on oncogenesis & biology of disease, preclinical therapeutic research & highlights of ongoing clinical translation. Register Now! https://t.co/bn7tcPelLA
#LCSM
0
3
19
Looking forward to @IASLC #SCLC23! So much fun to help organise this meeting together with superstars @Sutherland_Lab, @DMacPhersonLab, Kathryn Simpson and @charlesrudin. So many amazing talks with the latest advances in #SCLC. Limited spots! - register now! 👇
Join us for #SCLC23! A meeting focused on preclinical/clinical advances in #SCLC research, including basic research on oncogenesis & biology of disease, preclinical therapeutic research & highlights of ongoing clinical translation. Register Now! https://t.co/bn7tcPelLA
#LCSM
1
6
25