Centre for Computational Evolutionary Morphometry
@CCEM_ucph
Followers
117
Following
23
Media
15
Statuses
66
Evolutionary Biologists and Computer Scientists collaborate to address foundational questions on the Tree of Life, thanks to VILLUM and Novo Nordisk Foundations
Joined March 2022
Du @th_trad and I have a new preprint on arxiv: Parallel transport on matrix manifolds and Exponential Action https://t.co/zMpb1CDhpz
1
2
10
Hyperiax 2.0 is out! @marcustellx and the rest of the Hyperiax team have done an amazing job at revising the interface and in using Jax much more efficiently for tree computations. Hyperiax now traverses trees with >500 mill. nodes in <50 ms.
1
4
10
Kicking off Geometric Morphometrics workshop at @UCPH_Research and @domibujnak teaching 3D landmarking!!! 🐺
0
4
16
Score matching for bridges can be learned without time-reversal https://t.co/ezYczaQh3j w/ @thelibbybaker @MoritzSchauer We learn grad log p(t,x; T,y) for a target y directly without reversing time by combining score matching with adjoint diffusions (Milstein 2004) that give the
1
14
49
We welcome our new postdoc @RicardoAranda44, cosupervised with @ras_nielsen as part of our @VILLUMFONDEN project Stochastic Morphometry. Ricardo will study bird beak evolution (like in this kākāpō) using methods developed by our @CCEM_ucph team.
0
1
8
@thelibbybaker @gefanyang @Michael_Lind_S
@MoritzSchauer @MeulenFrank @mortenakhoej
@FrozakenDk Steven Evans @Huelsenbec20001 Nicklas Boserup, Chao Zhang, Thorben Pieper, Ricardo Ely, Jacky Li, Marc Corstanje @stefanhsommer
@ras_nielsen Christy Hipsley, Mads Nielsen
0
0
3
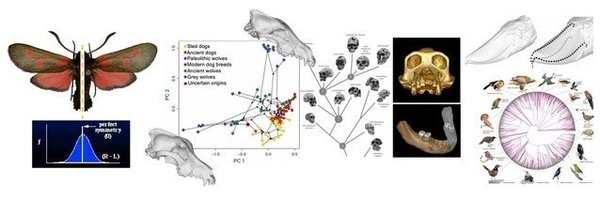
This week CCEM hosted a 3-day workshop featuring talks on phylogenetic and morphological analytics and stochastic processes as well as discussions with colleagues from UC Berkeley, Chalmers Uni., VU Amsterdam and Uni. of Delft. Thank you to all speakers and the VILLUM Foundation!
1
4
12
Second-order differential operators, stochastic differential equations and Brownian motions on embedded manifolds w/ Du Nguyen @th_trad
1
10
88
Conditioning shape processes is not restricted to finite configurations of landmarks - we can lift to infinite dimensions and condition nonlinear processes using Doob's h-transform, talk today by @thelibbybaker at CIRM Luminy
1
2
5
Simulating infinite-dimensional nonlinear diffusion bridges w/ @gefanyang @thelibbybaker @Michael_Lind_S Christy Hipsley We merge score matching with neural operators to simulate infinite dimensional non-linear diffusion bridges, e.g. bridges between shapes, ... 1/
1
16
95
@MeulenFrank @MoritzSchauer Coming soon will be other examples from the above references, and later on SDE cases and inference in SDE models for shapes. A major focus at @CCEM_ucph is to make inference in infinite dimensional models of shape variation along phylogenetic trees possible. The above notebooks
0
1
1
Our notebook Upwaards LDDMM utilizing the Hyperiax framework https://t.co/aJICl3L6AA… ..., is used here to estimate the ancestral shape (in blue) and two intermediate shapes (in green) for four different butterfly species belonging to Papilionoidea. @CCEM_ucph
0
3
10
Our notebook, https://t.co/aJICl3L6AA ..., implements a fast estimation of shape in inner nodes in a phylogenetic tree. Hyperiax is used here to estimate the intermediate shape(in blue) between two different species of butterflies! @CCEM_ucph
0
3
6
Our notebook https://t.co/xZWLqK1MaJ implements fast estimation of inner nodes of a phylogenetic tree - a simple application of our new tree traversal framework Hyperiax! We also show how Hyperiax can be used for fast simulation of trees under a Brownian Motion model. @CCEM_ucph
github.com
Tree traversals using JAX. Contribute to ComputationalEvolutionaryMorphometry/hyperiax development by creating an account on GitHub.
0
3
8
Hyperiax: Tree traversals using JAX https://t.co/Mo3ROHZrDH Developed by the team at @CCEM_ucph, initially for applications in phylogenetic analysis of morphological data, but Hyperiax has evolved into a general tree traversal, message passing and edge/node computation framework.
1
11
29
CCEM's PhD student Michael Severinsen is currently on a research stay at UC Berkeley - pictured here in front of dog skulls affected by artifical selection in California Academy of Sciences which is one of the foci of the research during his stay #VillumFoundation
0
0
7
Come join us for a postdoc in Copenhagen aimed at developing computational methods to map genotype-phenotype interactions in birds.
candidate.hr-manager.net
1
38
54
CCEM Annual Workshop 2023 was concluded by inspiring talks by Kyle Copas on GBIF – Global Biodiversity Information Facility, by Guojie Zhang on the B10K bird genome project and by Gavin Thomas on the Mark My Bird project. Thank you to all for great discussions! #VillumFoundation
0
3
7
First day of CCEM's annual workshop 2023 featured engaging discussions and six exciting talks on phenotypic and morphological analyses, evolutionary modelling of shape data, phylogenetic root estimation and species tree inference. #VillumFoundation
0
2
5