Bhandoola_Lab
@Bhandoola_Lab
Followers
21
Following
3
Media
0
Statuses
18
Our colleagues will of course decide whether the new NK classification proposed by @EricVivier1 is useful. What their decision, I do think a classification that makes sense of both mouse and NK subsets would be very useful to the field.
0
0
0
Our contribution was to show that mouse NK cells develop by 2 pathways, and that the transcriptional signatures that defined these 2 mouse NK subsets also distinguished human NK subsets defined by CD56.
0
0
0
Glad to have contributed to the work by Rebuffet, @EricVivier1 and colleagues, reposted below.
0
0
0
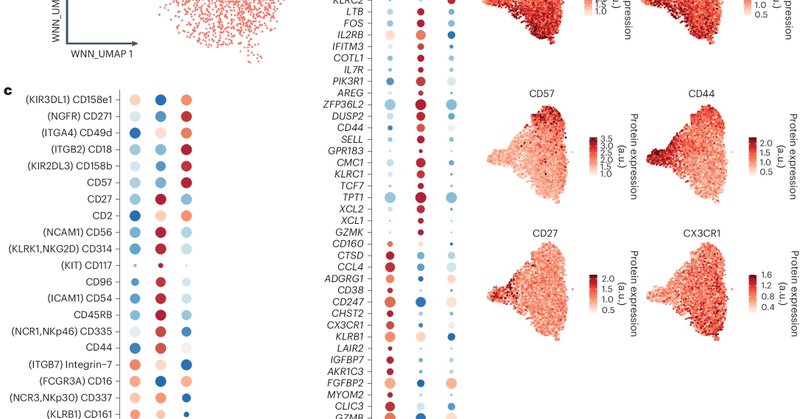
Vivier and colleagues integrated scRNA-seq and CITE-seq data to provide a resource of human NK cell heterogeneity in healthy tissues and in the tumor setting. Read it here: https://t.co/fTaQp3LNxC
https://t.co/yZrLSXRhhl
nature.com
Nature Immunology - Single-cell technologies have unveiled a complex understanding of NK cells that has led to variations in nomenclature and inconsistencies across the scientific literature. Here,...
1
14
91
And here are the links to the two new papers in Nature Immunology. From us and our collaborators @SimonGrassmann, @SunLab_Official, @BelkaidLab
https://t.co/ZaxtS0G5Sa From the lab of Bendelac and McDonald: https://t.co/ZYbuf9mfrJ
0
2
7
And here is the thoughtful N&V from Emma Patey & @bjorkstrom_lab
https://t.co/GCseoLjcc4
0
1
5
Such a bittersweet surprise to be back to back in Nature Immunology with Albert. To my thinking, our two papers are very complementary.
0
0
0
The original papers are here, one from our lab, one from the lab of Albert Bendelac. https://t.co/WWLyIlhDsC
https://t.co/6nFFQXtOha
0
0
1
Great summary from Emma Patey & @bjorkstrom_lab about the 2 recent papers studying NK cell developmental origins from the late Bendelac's lab & the Bhandoola's lab: https://t.co/GvLLvBDMrm ILCs with more nuance! Orginal studies: https://t.co/MFfl2XBFlG
https://t.co/DBA6g0l0RJ
0
3
8
Bhandoola and colleagues describe the existence of two developmental pathways that give rise to functionally distinct NK cell subsets. Read it here: https://t.co/seVYU7nx0t
https://t.co/HHZOT9yRMF
1
14
44
The CCR media team has a succinct outline of this latest work from our lab: https://t.co/pp6CPWHjdt
0
2
3
Online now: Transcription factor Tox2 is required for metabolic adaptation and tissue residency of ILC3 in the gut
0
20
77
https://t.co/rruCpomL41 Other recent work from our lab. Stay tuned for more.
science.org
Klf6 is required in thymic epithelial cells for normal thymic development and the establishment of T cell tolerance.
0
0
1
And here's the link to the paper itself, below. https://t.co/aIehlInOeC
nature.com
Nature - We demonstrate the role of the ligand-gated purinergic ion channel P2RX4 in maintaining mouse plasma cells in their bone marrow niche.
0
0
1
Bone marrow plasma cells require P2RX4 to sense extracellular ATP https://t.co/mZIT0bi4fB The link is to a web-readable version of our lab's first paper on plasma cells. From a very fun collaboration between our lab at the National Cancer Institute and David Allman's lab at Penn.
0
0
1