Benjamin Minch
@Benjamin_Minch
Followers
137
Following
463
Media
10
Statuses
62
Ph.D. student at UMiami Rosenstiel studying ocean viruses.
Joined February 2022
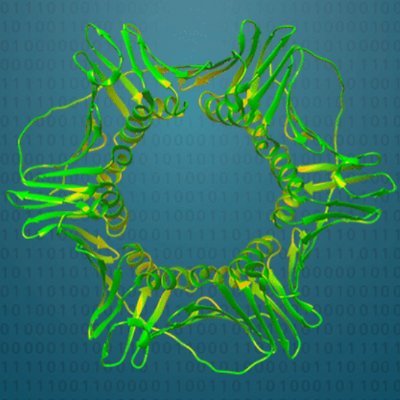
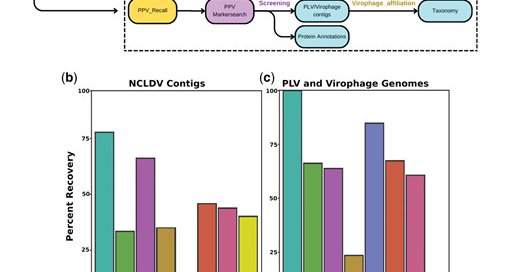
Are you interested in finding giant viruses, virophages, and PLVs inside your metagenomes? Struggling to figure out an easy way to do so? Look no further than BEREN, a one-stop-shop tool for recovery, taxonomy, and annotation of these viruses. https://t.co/OMCDlAGXcT
biorxiv.org
Viruses in the kingdom Bamfordvirae , specifically giant viruses (NCLDVs) in the phylum Nucleocytoviricota and smaller members in the Preplasmiviricota phylum, are widespread and important groups of...
3
14
55
🦠 Now published in Bioinformatics Advances: “BEREN: A bioinformatic tool for recovering giant viruses, polinton-like viruses, and virophages in metagenomic data.” Full article available: https://t.co/oImIhEPgui Authors include: @Benjamin_Minch
1
1
1
Excited to share our new paper introducing BEREN, a tool for identifying giant and other dsDNA viruses from metagenomic data — now online! Let BEREN help with your discovery-based reseach in viral metagenomics! https://t.co/H3DqmZBO0y
academic.oup.com
AbstractMotivation. Viruses in the kingdom Bamfordvirae, specifically giant viruses (NCLDVs) in the phylum Nucleocytoviricota and smaller members in the Pr
0
2
4
Created a fun phytoplankton-themed Oregon trail style game you can play on your terminal. Check it out https://t.co/i57B6nCnmt
github.com
A fun and educational Oregon trail style game about phytoplankton. - BenMinch/PhoticZone
0
0
0
We’re recruiting a PhD student (Fall 2026) to study the ecology & evolution of aquatic giant viruses! Research spans lab experiment, field sampling, & bioinformatics. Background in microbiology/bioinforamtics encouraged. More information: https://t.co/bshyrMLtiW Please retweet!
monirlab.com
1
14
40
Scientists used bespoke computer software to identify the genomes of microbes in seawater samples – including 230 giant viruses previously unknown to science. @univmiami @Benjamin_Minch
https://t.co/L9oBdVcBt9
0
3
8
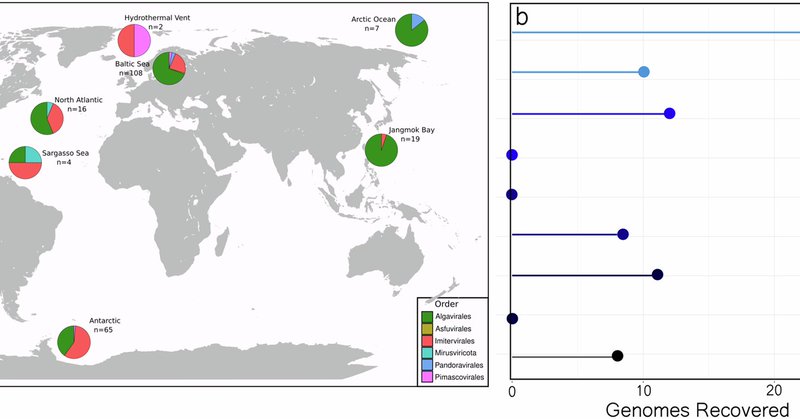
New paper out where we highlight many newly recovered giant viruses and find new functional potential encoded in their genomes! #giantvirus #Bioinformatics
https://t.co/lEK95lBUcc
nature.com
npj Viruses - Expansion of the genomic and functional diversity of global ocean giant viruses
0
12
35
Just put together a super easy-to-use and comprehensive pipeline for calculating various genome statistics in bulk (GC, length, codon bias, TNF, complexity, N50, num genes). https://t.co/VO6FSLzFGp
#bioinformatics #genomics
github.com
Contribute to BenMinch/genestat development by creating an account on GitHub.
0
0
2
🚨New @delCampoLab Preprint: A Single-cell Atlas of Coral Bleaching! 🪸🦠🔥. We show the microbial ecology of coral bleaching from the gene expression of host and symbiont cells to the shifts in the bacterial and protist communities within the holobiont!
researchsquare.com
The symbiosis between corals and dinoflagellate algae is disrupted by heat stress, leading to bleaching. Recurring bleaching events, driven by the climate crisis, are causing massive coral mortality...
1
20
40
What if we could put all tailed phages in a single, evolutionarily meaningful phylogeny? Check out the latest from the Aylward Lab (twitterless) and myself - we present a phylogenomic framework toward unifying phage diversity! 🧬🤝
biorxiv.org
Background Classifying viruses systematically has remained a key challenge of virology due to the absence of universal genes and vast genetic diversity of viruses. In particular, the most dominant...
0
7
20
BEREN is available on my gitlab
gitlab.com
A bioinformatic pipeline for identifying giant viruses and Preplasmiviricota viruses in metagenomic data.
0
0
2
BEREN: A bioinformatic tool for recovering Giant viruses, Polinton-like Viruses, and Virophages in metagenomic data
biorxiv.org
Viruses in the kingdom Bamfordvirae , specifically giant viruses (NCLDVs) in the phylum Nucleocytoviricota and smaller members in the Preplasmiviricota phylum, are widespread and important groups of...
0
5
18
Couldn't find a file to easily convert from PDB (protein data bank) IDs to actual protein information so I decided to make one. Hope someone finds it useful.
0
0
3
It's Thursday! Time for a new episode of #MattersMicrobial to be released! This time, the #QualityQuorum will hear from @giant_virus about giant viruses and how they interact with their hosts. Please spread the #GoodMicrobialWord. @ASMicrobiology
https://t.co/wOQCluAZQ0
1
3
16
Our latest collaborative research on giant viruses with @JedFuhrman lab on how evolutionary history dictates the temporal succession of marine giant viruses. Preprint out! @Benjamin_Minch
https://t.co/JccVG6KP0h
biorxiv.org
Nucleocytoplasmic Large DNA Viruses (NCLDVs, also called giant viruses) are widespread in marine systems and infect a broad range of microbial eukaryotes (protists). Recent biogeographic work has...
0
8
42
We also highlight a stark functional divide between the two most common types of Giant virus in the ocean, possibly hinting at different infection strategies. (4/4)
0
0
0
Using this pipeline we discovered >230 new genomes with cool functions not found in previous Giant Viruses. (3/4)
1
0
0
We introduce a streamlined pipeline for the recovery of Giant Viruses from metagenomic data ( https://t.co/GSw9RblQov) (2/4) #Bioinformatics
1
0
0
Go check out our latest preprint where we discover a bunch of new Giant Viruses encoding new functions all over the ocean. (1/4) 🧵 Expansion of the genomic and functional diversity of global ocean giant viruses https://t.co/Wfx3PK1CrK
#giantvirus
1
1
7
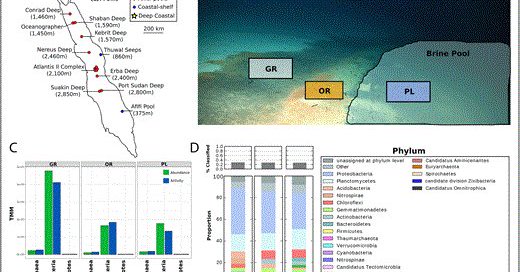
Active viral ecology of deep sea brine pool in the Gulf of Aqaba. Latest from our lab, and great collaboration with UMiami Marine Geoscience department. Spearheaded by Monir lab PhD student @Benjamin_Minch! https://t.co/HBdgkqX59v
academic.oup.com
Abstract. Deep-sea brine pools represent rare, extreme environments, providing unique insight into the limits of life on Earth, and by analogy, the plausib
2
7
38