Barkai Lab
@BarkaiLab
Followers
2K
Following
768
Media
123
Statuses
373
Systems Biology Lab @ 🔬Weizmann Institute of Science. # Understanding design principles of biological circuits.
Israel
Joined September 2017
A preprint‼️that's bound to ruffle some 🪶 "Widespread DNA off-targeting confounds studies of RNA chromatin occupancy" led by our @MicahGoldrich and Louis Delhaye from @pieter_mestdagh. TL;DR we show that many of lncRNA chromatin occupancy maps are flawed🧵 https://t.co/eCq6nORxQm
biorxiv.org
The importance of long noncoding RNA (lncRNA) functions is recognized across biological systems, but their modes of action remain poorly understood. One mechanism proposed to be particularly common...
1
14
44
Our analysis of the data suggests that there may not be a universal answer. Rather, we favor cross-validating the conclusions across definitions (e.g, thresholds, peak definition), and against orthogonal biological datasets.
0
0
2
There are pros and cons to focusing on top-bound targets vs. including all significant peaks. Can we assume that binding signals are proportional to binding affinities? And what would be the threshold for 'top-bound'?
1
0
0
Here is one example from our analysis: Ste12 is predicted to bind at more than 3,300 promoters, yet only 34 pass the top-bound threshold. Most of these 34 targets are induced by mating pheromones, consistent with Ste12 being the known regulator of the yeast mating response.
1
0
0
A recent study reported a surprisingly low overlap between TF binding and regulatory targets. But this may depend on how binding sites are defined, as we report; Should one include all statistically significant peaks or focus on top-bound targets? https://t.co/sx7fkoxGnn
biorxiv.org
A recent study that systematically mapped genomic bindings and regulatory effects of transcription factors (TFs) reported a surprisingly low overlap between TF binding and regulatory targets in...
2
8
51
Design principles of transcription factors with intrinsically disordered regions @eLife @BarkaiLab
https://t.co/gv1weMyUSi We study how IDRs allow TFs to efficiently locate DNA targets while maintaining strong binding. 1/3
elifesciences.org
Intrinsically disordered regions in transcription factors enhance both target binding probability and search efficiency.
1
5
13
Why are transcription factors disordered? Come join us in Oxford as a postdoc and we'll find out! Help publish 3 mature projects, AND develop cool new single molecule fluorescence binding assays! biophysics transcription protein:DNA interactions https://t.co/uxDhTEv1eL
2
7
27
10% of human transcription factors are oligomeric. Stoichiometry can be dialled to modulate transcription. We show CREB searches DNA mixtures as a dimer, and suggest this might be common amongst other members of the bZIP family which fold on binding DNA. https://t.co/4UZWB9Odaw
biorxiv.org
The operation of Eukaryotic transcription factors remains enigmatic. Cyclic AMP-responsive element-binding protein (CREB) is a member of the basic zipper family, a superfamily of transcription...
2
6
47
🧵 1/ New preprint! How do transcription factors (TFs) use intrinsically disordered regions (IDRs) to find their target sites? https://t.co/ugZ1oQV4gu
#TranscriptionFactors #IDPs #SingleMolecule #Biophysics
biorxiv.org
Transcription factors (TFs) regulate gene expression by binding specific DNA motifs, yet only a fraction of putative sites is occupied in vivo . Intrinsically disordered regions (IDRs) have emerged...
2
15
56
Now out on @NAR_Open
https://t.co/3P8u7VHOsX
academic.oup.com
Abstract. DNA-binding domains (DBDs) within transcription factors (TFs) recognize short sequence motifs that are highly abundant in genomes. In vivo, TFs b
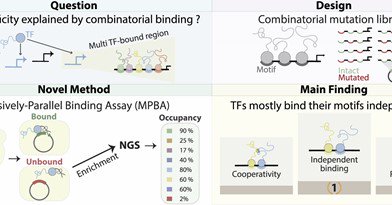
New 📄 by @tamar_jana + @SagieBr We introduce MPBA, a novel method allowing parallel measurement of TF binding to thousands of designed DNA sequences within cells. We use it to answer a fundamental question - how prevalent is TF binding cooperativity. 1/9
0
8
17
(13/13) This study redefines our understanding of coactivator recruitment, highlighting the active role of Med15 in transcriptional regulation. Could this lead to new insights into gene expression modulation and potential therapeutic targets? Stay tuned!
1
0
3
(12/13) We propose a two-step recognition model in which the Med15 first locates its target promoters and then is stabilized by the interactions with other TFs.
1
0
2
(11/13) While Med15 and the DBD do not localize alone to Med15 native promoters in this strain, their fusion rescued the binding, indicating that Med15 can independently find its target locations.
1
0
1
(10/13) To test this, we mapped the binding locations of our Med15-DBD fusion in strain co-deleted of 12 Med15-recruiting TFs.
1
0
1
(9/13) We took it a step further by directly fusing Med15 to DBDs. This fusion redirected the DBD binding away from the newly acquired minimal TF targets toward fuzzy nucleosome promoters, suggesting that Med15 itself has an inherent preference for these locations.
1
0
1
(8/13) Is Med15 even recruited to these promoters? To understand this further, we mapped its binding locations in cells expressing our minimal TFs. With agreement to the expression profiles, Med15 recruitment occurred primarily at induced promoters with fuzzy nucleosomes.
1
0
1
(7/13) Surprisingly, the newly acquired targets of the DBD-ADs were not induced—only those also occupied by the full TFs showed increased expression. These shared and induced targets were characterized by fuzzy promoter architecture.
1
0
1
(6/13) By mapping the binding locations of the DBD-ADs, we discovered that they acquire new binding sites not occupied by the full TFs. Interestingly, the identity of the fused AD also influenced the binding preferences of the minimal TF.
1
0
1
(5/13) Expression profiling revealed that the full-length TFs are generally more effective at gene induction than the DBD-AD fusions.
1
0
1
(4/13) Accurate measures of genome-wide transcription are not trivial. For this, we used synthetic promoter libraries, expressing our minimal TFs at increasing levels (up to 40 different strains for each TF).
1
0
1