AtlasXomics
@AtlasXomics
Followers
616
Following
42
Media
34
Statuses
66
Accelerating Spatial Multi-Omics
New Haven, CT
Joined November 2020
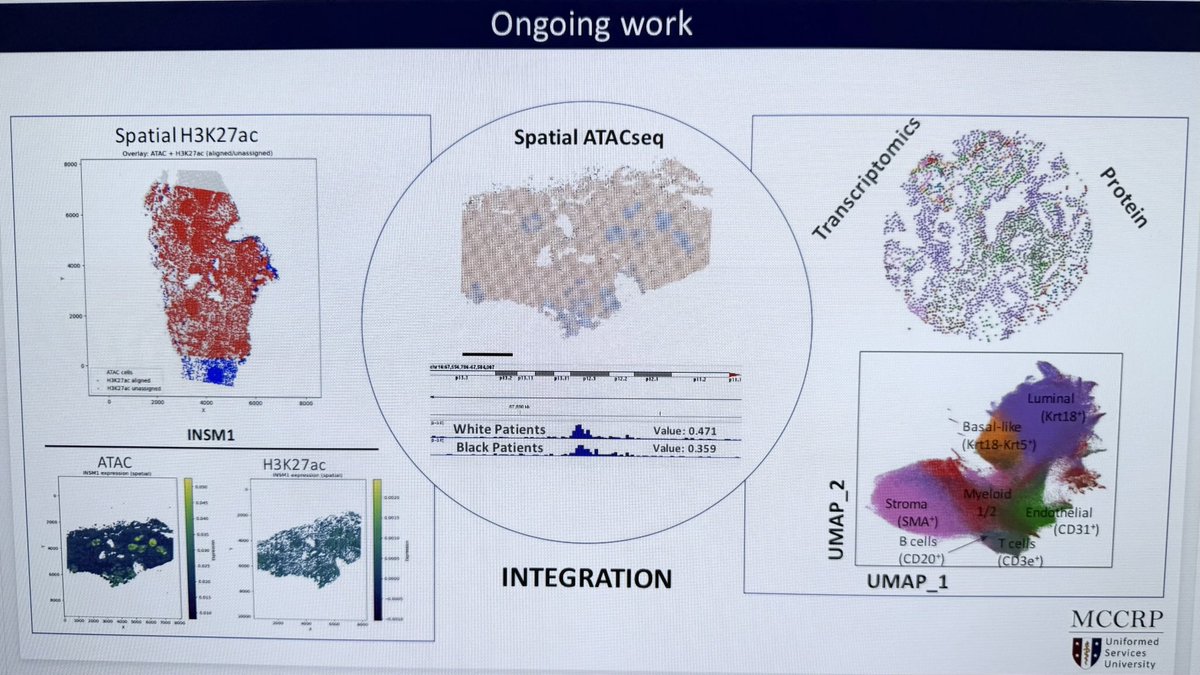
🚨 New preprint! Dr. Marco Gallo’s lab used AtlasXomics Spatial ATAC‑seq to map chromatin in GBM and found nested epigenomic niches that drive cell‑state plasticity. Dive in: #Glioblastoma #CancerResearch #Epigenetics.
biorxiv.org
IDH -wildtype glioblastoma (GBM) is an aggressive brain tumor with poor survival and few therapeutic options. Transcriptionally-defined cell states coexist in GBM and occupy defined regions of the...
0
1
7
Exciting News! FlowGel is now a routine part of YCGA’s toolkit, powering spatial epigenomics for Yale labs. Huge thanks to Bill Renock and Bony De Kumar for guiding the rollout and keeping the data flowing.🚀 #Epigenetics #Epigenomics #SpatialOmics
1
2
12
On a roll! 🎉 @YanxiangDeng’s lab + @zhouwanding drop a new preprint extending #DBiTseq to co-map DNA methylation 🧬 & RNA in the same tissue slice. Kudos @ChinNLee2021 & Hongxiang Fu!.
DNA methylation goes spatial! Introducing Spatial-DMT: a technology that co-profiles DNA methylation and transcriptome in the same tissue section. A fantastic collaboration with @zhouwanding lab. Kudos to Chin Nien Lee @ChinNLee2021 and Hongxiang Fu!
0
0
7
🧬 Kudos to The Deng Lab @PennMedicine!.New Nat Comms: spatial FFPE-ATAC for DBiT-seq maps chromatin in archived brain, thymus & melanoma (10 µm) with AtlasXomics FlowGel featured. We’re working to bring this to researchers. Reach out to learn more!.#SpatialOmics #Epigenetics.
Our spatial FFPE-ATAC-seq is now published in @NatureComms! With extensive optimizations, it performs well across diverse tissues, unlocking the vast FFPE archive for spatial epigenomics. Big congrats to Pengfei Guo @pengfeo, Yufan Chen, and the team!
0
1
7
What a fantastic talk by @BeatrizGerFalc and @EllisLab_ at the Cedars-Sinai Single Cell and Spatial Omics Symposium! We’re proud to enable such exciting insights.
@BeatrizGerFalc representing @EllisLab_ discussing our spatial epigenomic analysis in #prostatecancer #spatialAtacseq. @PCF_Science @UrologySBUR @DrJasPlummer @LGMartelotto @RongFan8 @AtlasXomics
0
2
11
New preprint from Dr. Xusheng Wang (UTHSC)! Using AtlasXomics’ spatial ATAC‑seq kit, the team mapped chromatin shifts in six‑month‑old 5×FAD Alzheimer’s mice and uncovered microglial TREM2 & VISTA hotspots that fuel neuro‑inflammation. Read ➡️
biorxiv.org
Abnormal epigenetic modifications, including altered chromatin accessibility, have been implicated in the development and progression of Alzheimer’s disease (AD). In this study, we applied spatially...
0
0
9
Want to run spatial epigenomics in your lab? Our DBiT-seq kit makes it easy. Watch how it works! #epigenetics #spatialbiology
0
1
4
Our CEO, Ken Wang, takes the stage at Nvidia’s GTC conference, sharing how our advanced computational pipelines are shaping the future of spatial epigenomics. Stay tuned for more innovative breakthroughs! #GTC #nvidia #epigenetics
0
2
10
Unlock the hidden secrets of tissues with AtlasXomics! Discover how our revolutionary DBiT-seq enables spatial epigenomics. #Epigenetics #Genomics #SpatialBiology
0
2
9
Discover how we’re turning the tide on tumor heterogeneity. Watch our new video to see how Atlasxomics’ cutting‐edge spatial epigenomic tools are powering breakthroughs in cancer research. #CancerResearch #SpatialEpigenomics
0
1
6
Exciting to see our spatial ATAC-seq assay featured in @PAINthejournal! 🔬✨ This study by Urzula Franco-Enzástiga unveils the epigenomic landscape of the human dorsal root ganglion, shedding light on sex differences in nociceptive gene regulation. #SpatialEpigenomics #ATACseq.
0
1
6
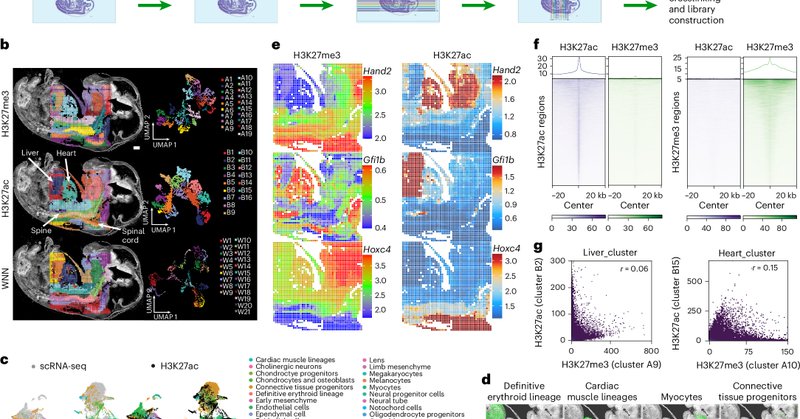
RT @DengYanxiang: Our spatial multi-omics technology that co-profiles five modalities is now published in Nature Methods @naturemethods! Hu….
nature.com
Nature Methods - Spatial-Mux-seq offers a multimodal spatial platform capable of profiling multiple molecular modalities, including the transcriptome, chromatin accessibility, histone modifications...
0
63
0
AtlasXomics is thrilled to see our collaborators at @LatchBio accelerate peak calling with a GPU-based MACS rewrite—achieving roughly 15X faster performance! This breakthrough significantly speeds up single-cell and spatial assays, enabling quicker epigenetic insights and.
Peak calling is important for epigenetics analysis but current tools aren't keeping pace with data generation. We're releasing a GPU implementation with ~15X speed up on industry workloads. A breakdown of the algorithm steps + benchmarks (1\N):
0
4
10
Cheers to a year of breakthroughs at Atlasxomics—and an even more innovative 2025 ahead! Together, let’s push the boundaries of discovery. #epigenetics #genomics
0
2
11
RT @RongFan8: Thrilled to share Perturb-DBiT — spatial unbiased in vivo perturb-seq with genome-scale CRISPR libraries! Hope you enjoy read….
biorxiv.org
Perturb-seq enabled the profiling of transcriptional effects of genetic perturbations in single cells but lacks the ability to examine the impact on tissue environments. We present Perturb-DBiT for...
0
61
0