Sukrit Singh
@sukritsingh92
Followers
1K
Following
51K
Media
487
Statuses
3K
Biophysicist|@DamonRunyon QB + @theNCI K99/R00 Fellow @MSKCancerCenter w @jchodera | @foldingathome|PhD w @drGregBowman|resistance, dynamics, dogs 🇮🇳🇸🇬🇺🇸
nyc
Joined June 2016
I'm on the job market looking for faculty positions! If you want to talk about protein dynamics, precision medicine, drug resistance, and/or puppy photos, reach out!
0
14
65
RT @kannandelhi: Ravikant Singh, 64, passes away. Knew him since 1987 when he was in Times of India and I was in Indian Express. We cover….
0
2
0
RT @ddsportschannel: The world of sports journalism has lost a true icon. Ravi Kant Singh, aged 64, passed away on Thursday, leaving behind….
0
4
0
RT @manishbatavia: Shocked to learn of the passing away of Ravi Kant Singh who I worked with on so many broadcasts over the last 9 years. H….
0
1
0
Now published in J Phys Chem B! Check out our work showing that simulations predict the impact of distal mutations on kinase-inhibitor binding, and our experimental NanoBRET dataset of 94 kinase mutations that provide a benchmark for future methods. Link:
New preprint! We prospectively evaluate structure-based methods and show how well they classify kinase mutations as resistant or sensitizing to inhibitors when compared to our NanoBRET assay. Alchemical methods are well-suited to modeling distal mutants!
0
3
14
RT @XuhuiHuangChem: Exciting news! Our TS-DAR method is now published in @NatureComms! 🎉.TS-DAR identifies transition states across multipl….
0
18
0
RT @foldingathome: Too many requests for money this Giving Tuesday?. Consider giving your spare CPU/GPU cycles to advance biomedical resear….
0
10
0
Shoutout to citizen-scientists @foldingathome where we ran work units to retrospectively analyze the "really bad" predictions, and identify ways we can improve them!.
0
1
3
New preprint! We prospectively evaluate structure-based methods and show how well they classify kinase mutations as resistant or sensitizing to inhibitors when compared to our NanoBRET assay. Alchemical methods are well-suited to modeling distal mutants!
Prospective evaluation of structure-based simulations reveal their ability to predict the impact of kinase mutations on inhibitor binding #biorxiv_biophys.
1
5
21
RT @JoshRackers: We can accelerate molecular conformational ensemble sampling with generative AI!. Walk-Jump Sampling smoothes out the dist….
0
13
0
RT @mia_rosenfeld: wow, what an amazing and massively useful guide to large parallel simulations from @sukritsingh92 and @sonyahans !!!! 🤩💻.
0
1
0
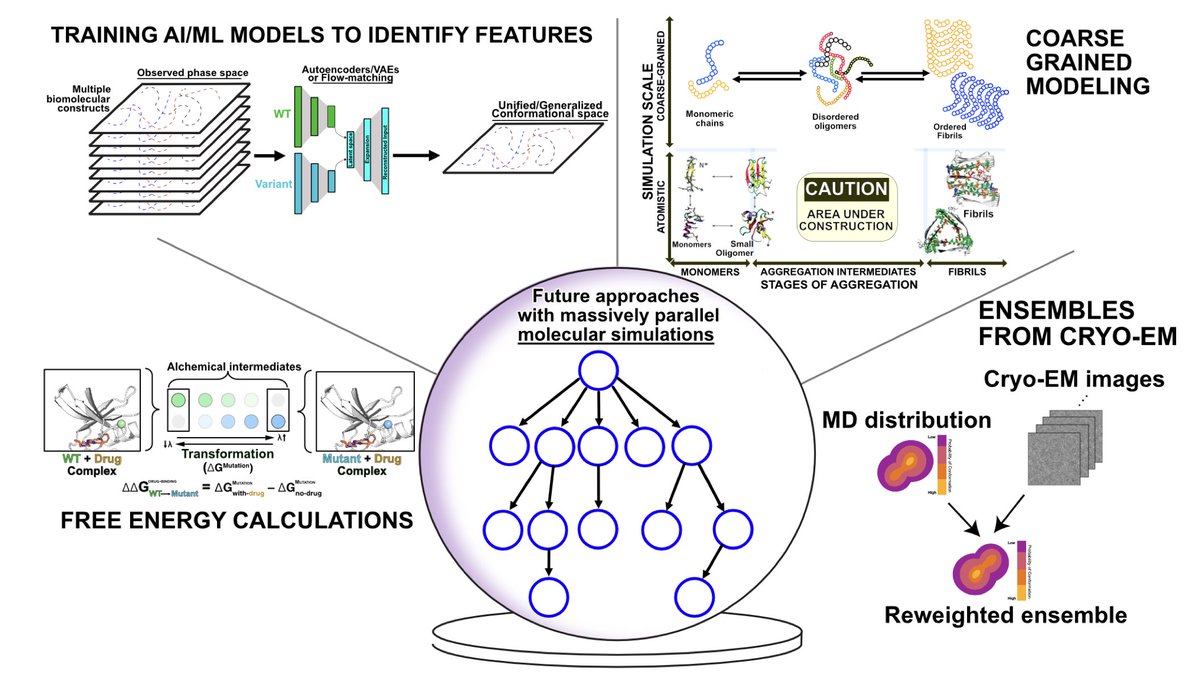
Excited to share my and @sonyahans's little guide to large parallel simulations (and what you can do with them) in preprint form!.
Running and Analyzing Massively Parallel Molecular Simulations. 1. This article provides a comprehensive guide on generating and analyzing massively parallel molecular dynamics (MD) simulations to explore protein conformational landscapes and their role in biological processes.
0
13
33
RT @BiologyAIDaily: Running and Analyzing Massively Parallel Molecular Simulations. 1. This article provides a comprehensive guide on gener….
0
13
0
RT @sonyahans: The Conference for Undergraduate Women and Gender Minorities in Physics (CU*iP) application deadline is Oct 23rd! NYU is one….
aps.org
The 2026 CU*iP will be held in January. We welcome participation from all undergraduate physics students interested in building a more inclusive physics community.
0
4
0
RT @MarkHahnel: Big shout out to those researchers who made their data available in PDB over the years. The Chemistry Nobel Prize winners t….
0
212
0
RT @RolandDunbrack: Why Alphafold succeeded - the best large, curated, cleaned training data set is the PDB. Thank Helen Berman for that.
0
25
0
RT @ivyzhang__: Registration is now open for the next CADD GRS, taking place at the University of Southern Maine July 12-13, 2025! . Great….
0
7
0
RT @MSKEducation: We want to thank @MSKCancerCenter Communication for helping us bring the stories behind our scientists and giving us an i….
0
4
0
RT @gonenlab: Folks please remember, when looking for a lab to join, always talk with current *and* past trainees. Joining a lab that nurt….
0
12
0