Gajendra Raghava
@raghavagps
Followers
519
Following
23
Media
13
Statuses
275
Professor & Head Centre for Computational Biology, IIIT, Delhi
New Delhi, India
Joined April 2010
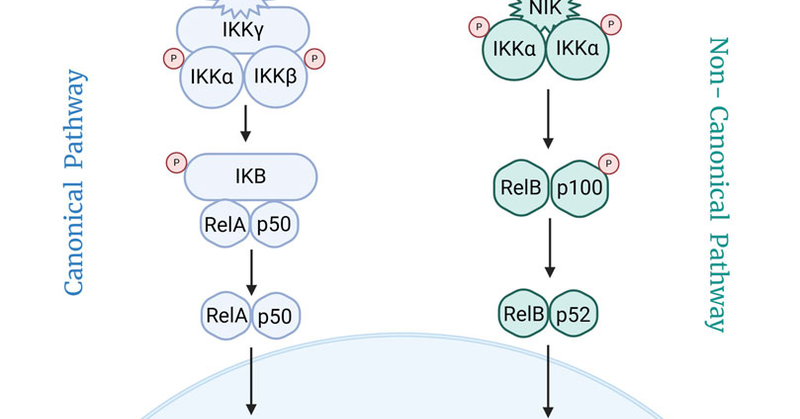
🧬 We present NFκBIn, a machine learning-based tool for identifying inhibitors of the TNF-α-induced NF-κB pathway — a key target in inflammatory diseases and cancer. 🌐 Try the webserver: 💻 Access the paper:
frontiersin.org
IntroductionNuclear Factor kappa B (NF-κB) is a transcription factor whose upregulation is associated in chronic inflammatory diseases, including rheumatoid ...
0
0
0
We present HAIRpred – a novel AI-based tool for predicting Human Antibody Interacting Residues in antigens. First time we developed method for predicting B-cell epitope for human host, existing methods are not host specific. 📄 Publication:
onlinelibrary.wiley.com
In the past, several methods have been developed for predicting conformational B-cell epitopes in antigens that are not specific to any host. Our primary analysis of antibody–antigen complexes...
0
0
0
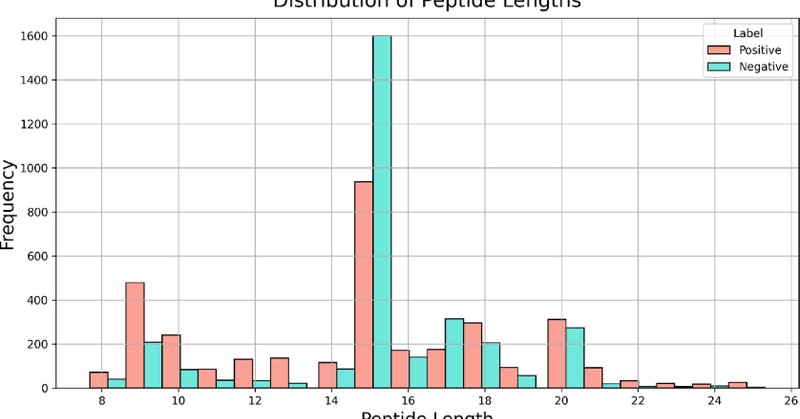
We present NTxPred2, an advanced AI-based tool for predicting neurotoxic peptides and neurotoxins. Unlike earlier version NTXpred, NTxPred2 uses separate models for peptides and proteins. 🔗 Webserver: .📄 Publication:
onlinelibrary.wiley.com
The accurate prediction of neurotoxicity in peptides and proteins is essential for the safety evaluation of therapeutic proteins and genetically modified (GM) organisms. Existing tools, including...
0
0
1
We are pleased to announce the launch of a new web server for immunoinformatics and vaccine design. This tool enables users to predict, design, and scan IL-2 inducing peptides. Paper link: Web Server
nature.com
Scientific Reports - In Silico tool for predicting, designing and scanning IL-2 inducing peptides
0
0
0
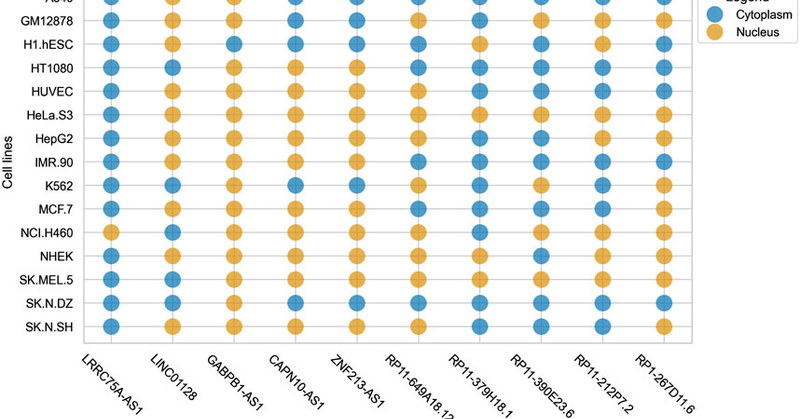
We are happy to announce new web service form group for predicting cytoplam associated lncRNA.
frontiersin.org
The function of long non-coding RNA (lncRNA) is largely determined by its specific location within a cell. Previous methods have used noisy datasets, includi...
0
0
0
We are happy to announce a new web server PPTstab for predicting and designing thermostable proteins with a desired melting temperature. Paper: Web : GitHub:
github.com
PPTStab: Designing of thermostable proteins with a desired melting temperature - raghavagps/pptstab
0
0
1
RT @ajitabhbhardwaj: Booked in Dec 2017. It’s April 2025, and still no possession at Godrej Nest, Sector 150 Noida. @GodrejProp — over 7 ye….
0
46
0
RT @IIITDelhi: Ph.D. Admissions (Monsoon 2025-26) are now open at IIIT-Delhi!. And, we have an exciting news for the Ph.D. aspirants. IIIT-….
0
9
0
Our group has developed a new server, EIPpred, for predicting, designing, and virtually scanning peptide-based inhibitors against bacteria (E. coli). Web Server: Paper:
nature.com
Scientific Reports - Prediction of inhibitory peptides against E.coli with desired MIC value
0
0
4
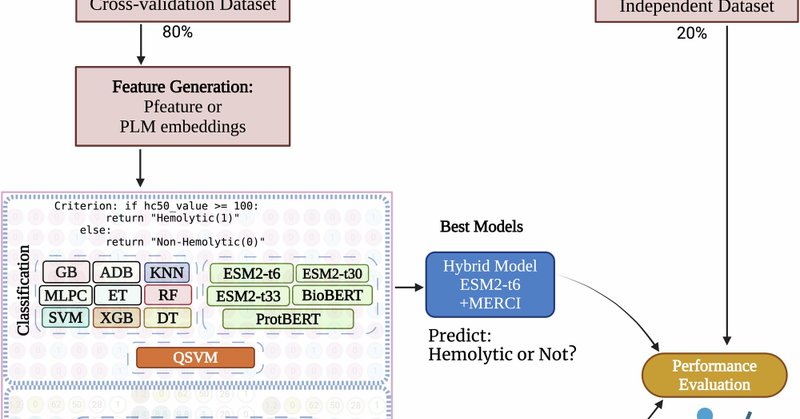
We are happy to announce our web server HemoPI2 an updated version of HemoPI. It is an advanced computational tool designed for predicting the hemolytic activity of peptides and estimating their hemolytic concentration (HC50). Paper link:
nature.com
Communications Biology - Developed models predict and quantify hemolytic activity of peptides. A hybrid model achieves AUROC of 0.921, regression models show R = 0.739. Implemented in...
0
0
3
Prediction of Plant Resistance Proteins Using Alignment‐Based and Alignment‐Free Approaches - Gahlot - PROTEOMICS - Wiley Online Library
analyticalsciencejournals.onlinelibrary.wiley.com
Plant disease resistance (PDR) proteins are critical in identifying plant pathogens. Predicting PDR protein is essential for understanding plant–pathogen interactions and developing strategies for...
0
0
1
We are happy to announce second generation software IFNepitope2 for predicting interferon-gamma inducing peptides for mouse and human host. Our previous method IFNepitope is widely used and cited by scientific community.
nature.com
Scientific Reports - A hybrid method for discovering interferon-gamma inducing peptides in human and mouse
0
0
3
Prediction of Anti‐Freezing Proteins From Their Evolutionary Profile - Kumar - PROTEOMICS - Wiley Online Library
analyticalsciencejournals.onlinelibrary.wiley.com
Prediction of antifreeze proteins (AFPs) holds significant importance due to their diverse applications in healthcare. An inherent limitation of current AFP prediction methods is their reliance on...
0
0
0
We are happy to announce new web server for predicting transcription factors
frontiersin.org
Transcription factors are essential DNA-binding proteins that regulate the transcription rate of several genes and control the expression of genes inside a c...
0
0
1