Nextstrain
@nextstrain
Followers
42K
Following
16
Media
2K
Statuses
3K
Real-time tracking of pathogen evolution. Also now on Mastodon: @[email protected]
Joined September 2016
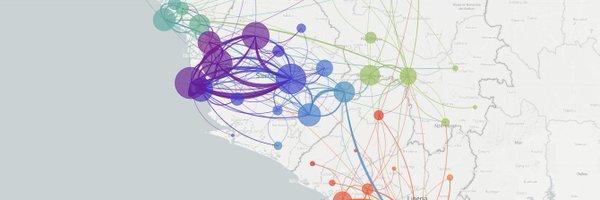
We've continued to track evolution and spread of H5N1 in cattle at as data has been shared by @USDA_APHIS. 1/3
4
43
84
This was an enormous amount of work. Congratulations to @ivan_aksamentov, @corneliusroemer and the @nextstrain team for pushing it across the finish line!. 8/9.
3
0
15
23I (BA.2.86), the parent of 23G (XBB.1.5.70) - XBB.1.5 - and the ancestor of 23H (HK.3) - EG.5 - have all been designated as Variants of Interest (VOI) by @WHO. More information by WHO TAG-VE can be seen here:. 9/9.
2
4
27
Additionally, we've updated the Nextclade datasets to appropriately call clade 23G, 23H, and 23I viruses at If you use #Nextclade in the browser, make sure to reload the tab from time to time so that you get the most recent datasets. 8/9
1
4
25