JASPAR database
@jaspar_db
Followers
331
Following
110
Media
6
Statuses
83
JASPAR is an #openaccess database of manually curated, non-redundant transcription factor (TF) binding profiles
Joined October 2017
Now you can query and access TF binding motifs from various releases of JASPAR database using BioPython without a MySQL connection. > pip install pyjaspar.> conda install pyjaspar -c bioconda. 💻code: 📘docs: cc @khanaziz84.
github.com
pyJASPAR: A Pythonic interface to JASPAR transcription factor motifs - asntech/pyjaspar
1
20
59
RT @GenomeBrowser: We are excited to announce the new JASPAR 2024 tracks for hg19, hg38, mm10, and mm39 which represent genome-wide predict….
0
12
0
RT @AMathelier: I am very excited to see the outcome of the extensive work provided by @elixirnorway Oslo to support JASPAR @jaspar_db in i….
0
2
0
RT @AMathelier: 🎉 Excited to announce the 10th release & 20th-anniversary update of JASPAR @jaspar_db , a leading resource in computational….
academic.oup.com
Abstract. JASPAR (https://jaspar.elixir.no/) is a widely-used open-access database presenting manually curated high-quality and non-redundant DNA-binding p
0
37
0
RT @AMathelier: It is always a great feeling to get a paper accepted on a Friday evening 🎉.JASPAR 2024 will soon be up and running for ever….
0
6
0
RT @mkuijjer: News article on our most recent publication 👇. Thanks @NCMMnews, and of course @tanja_belova for the great work ✨. https://t.c….
med.uio.no
The Kuijjer group present PORCUPINE, a computational tool for pinpointing drivers of cancer heterogeneity that may guide personalized cancer treatment.
0
2
0
RT @elixirnorway: #JobVacancy ELIXIR Norway, location @II_UiB, is hiring System #developers and System #administrators to work on our exci….
0
8
0
RT @BorisLenhard: Come join us! Two postdoc positions in regulatory genomics-computational and a (mostly) experimental-available in our gro….
0
39
0
RT @AMathelier: Great day with this amazing group of people! It was such an enjoyable full day of JASPAR @jaspar_db hackathon in sunny Oslo….
0
5
0
RT @AMathelier: @Francois_Parcy @JMFrancoZ @TrendsPlantSci @romain_bm Awesome work! Congrats @romain_bm @Francois_Parcy et al. Very valuabl….
0
1
0
RT @WyWyWa: After 18 years, the original JASPAR_DB paper has reached 2000 citations. Glad it has been useful. The….
0
3
0
RT @AMathelier: @jaspar_db The problem seems to be fixed. JASPAR is back up and running! @jaspar_db #JASPAR.
0
1
0
You can always query the motifs using the python package or R package
github.com
pyJASPAR: A Pythonic interface to JASPAR transcription factor motifs - asntech/pyjaspar
0
1
1
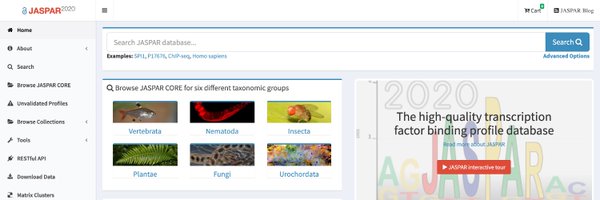
We've some glitches on our production web-server. Please use the development instance until further notice - We apologize for the inconvenience.
jaspar.uio.no
JASPAR is the largest open-access database of curated and non-redundant transcription factor (TF) binding profiles from six different taxonomic groups.
2
4
3
RT @GenomeBrowser: We are happy to announce the release of JASPAR Transcription Factor Binding Sites for hg19, hg38, mm10, and mm39. This r….
0
8
0
RT @khanaziz84: The @NAR_Open Database Issue 2022 is online with 87 new databases and 85 updates, including @jaspar_db .
0
7
0
RT @elixirnorway: Happy to share that we have added eight excellent Norwegian bioinformatics services to our service delivery plan with @EL….
0
2
0
RT @AMathelier: Great to share our new manuscript describing the latest update of JASPAR @jaspar_db. New TF binding profiles, familial bind….
academic.oup.com
Abstract. JASPAR (http://jaspar.genereg.net/) is an open-access database containing manually curated, non-redundant transcription factor (TF) binding profi
0
38
0
RT @WyWyWa: Excited that JASPAR 2022 is out today! It's a significant update, with new TF binding profiles, new enrichment analysis tools a….
0
21
0
RT @GenomeBrowser: Native @jaspar_db transcription factor tracks are now available for hg19 and hg38. JASPAR has long been a popular public….
0
8
0
RT @jaspar_db: Now you can query and access TF binding motifs from various releases of JASPAR database using BioPython without a MySQL conn….
github.com
pyJASPAR: A Pythonic interface to JASPAR transcription factor motifs - asntech/pyjaspar
0
20
0